Search Count: 259
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRUS |
 |
Organism: Rhodobacter capsulatus sb 1003
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRUS |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: CELL CYCLE |
 |
Cyclohexanone Dehydrogenase (Cdh) From Alicycliphilus Denitrificans K601 - Wildtype
Organism: Alicycliphilus denitrificans k601
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-02-14 Classification: FLAVOPROTEIN Ligands: FAD, GOL, SO4 |
 |
Cyclohexanone Dehydrogenase (Cdh) From Alicycliphilus Denitrificans K601 Complexed With Dehydrogenated Substrate Cyclohex-2-En-1-One - Inactive Mutant (Y195F)
Organism: Alicycliphilus denitrificans k601
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-02-14 Classification: FLAVOPROTEIN Ligands: FAD, A2Q, GOL, SO4 |
 |
Cyclohexanone Dehydrogenase (Cdh) From Alicycliphilus Denitrificans K601 Complexed With Dehydrogenated Substrate - W113A Mutant
Organism: Alicycliphilus denitrificans k601
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-14 Classification: FLAVOPROTEIN Ligands: FAD, GOL, PEG, A2Q, SO4 |
 |
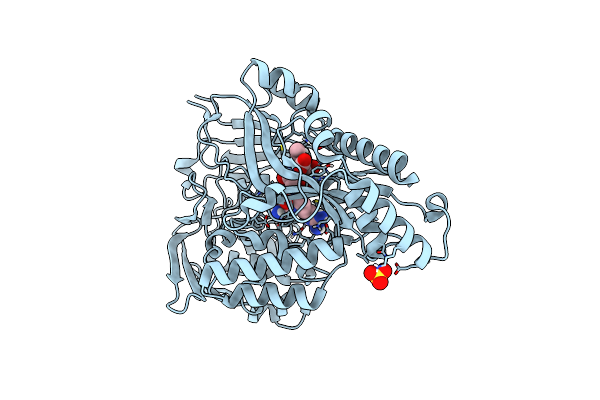
Structure Of Aap A Domain And B-Repeats (Residues 351-813) From Staphylococcus Epidermidis
Organism: Staphylococcus epidermidis rp62a
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-03 Classification: CELL ADHESION Ligands: CA, CL |
 |
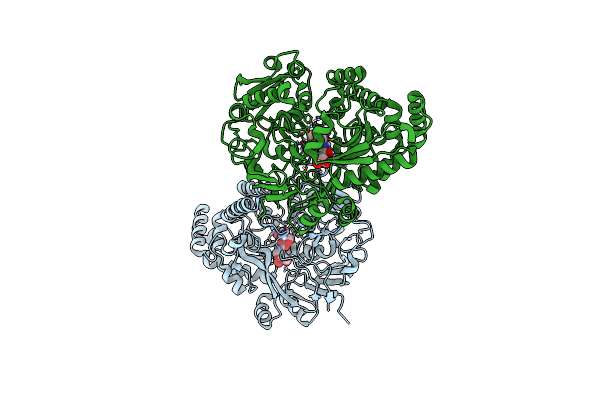
Crystal Structure Of The Peptide Binding Protein, Oppa, From Bacillus Subtilis In Complex With A Phre-Derived Pentapeptide
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
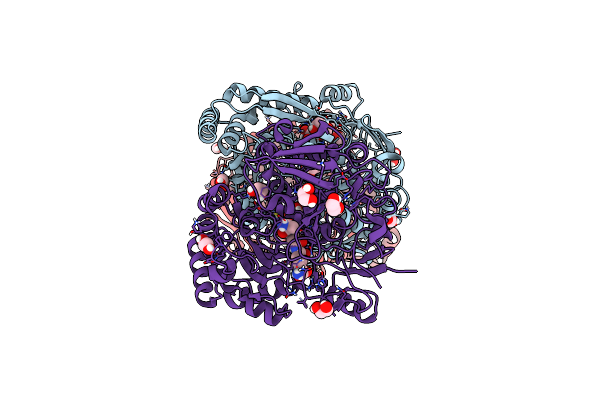
Crystal Structure Of The Peptide Binding Protein, Oppa, From Bacillus Subtilis In Complex With An Endogenous Tetrapeptide
Organism: Bacillus subtilis subsp. subtilis str. 168, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN |
 |
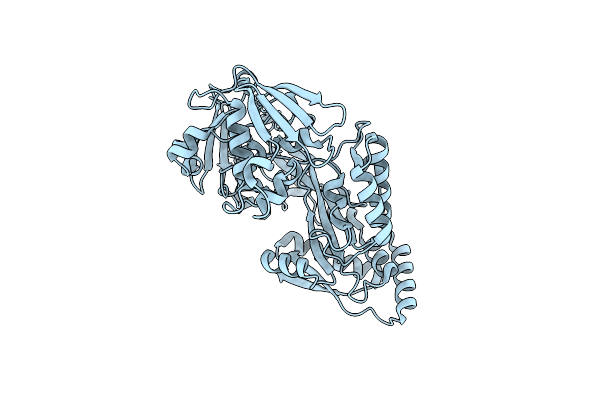
Crystal Structure Of The Peptide Binding Protein Dppe From Bacillus Subtilis In Complex With Murein Tripeptide
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: MHI, MG, EDO |
 |
Crystal Structure Of The Peptide Binding Protein Dppe From Bacillus Subtilis In The Unliganded State
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN |
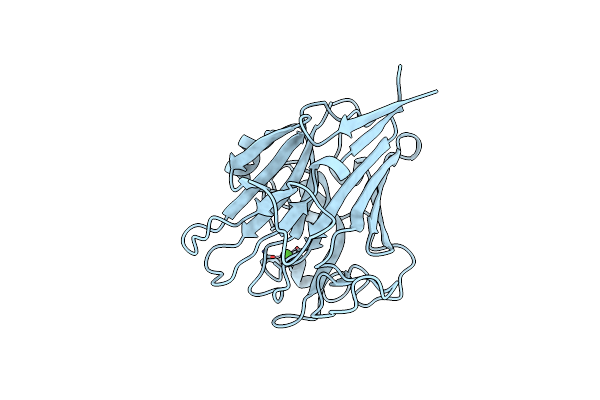 |
Structure Of Pls A-Domain (Residues 391-656; 513-518 Deletion Mutant) From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-11-09 Classification: CELL ADHESION Ligands: CA |
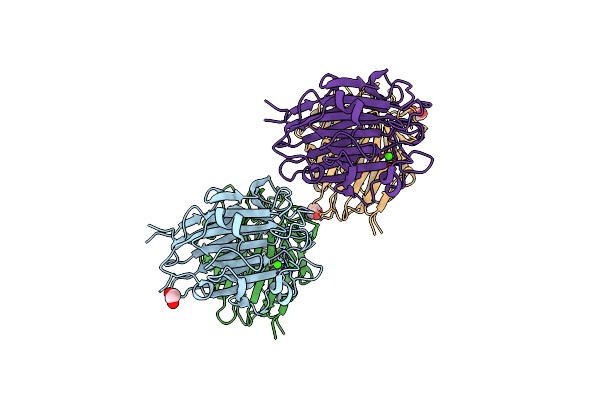 |
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-11-02 Classification: CELL ADHESION Ligands: CA, EDO |
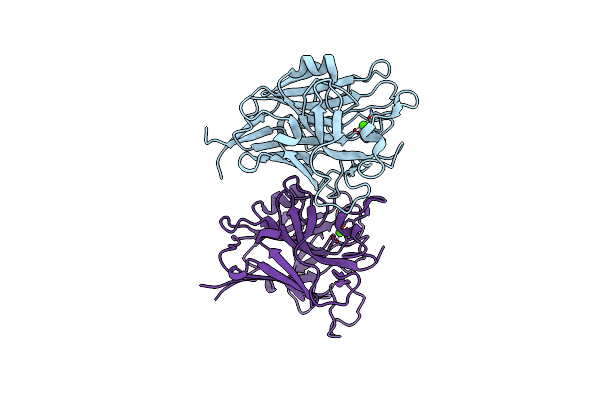 |
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2022-10-26 Classification: CELL ADHESION Ligands: CA |
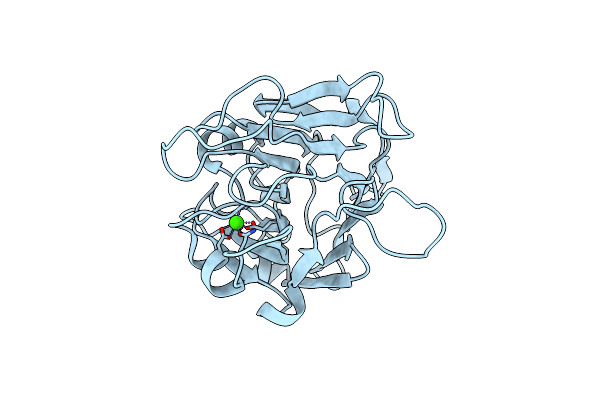 |
Structure Of Aap A-Domain (Residues 351-605) From Staphylococcus Epidermidis
Organism: Staphylococcus epidermidis (strain atcc 35984 / rp62a)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-10-19 Classification: CELL ADHESION Ligands: CA, CL |
 |
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2021-07-14 Classification: HYDROLASE |
 |
Organism: Trichobolus zukalii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Structure Of A-L-Araazi-Bound Mggh51 A-L-Arabinofuranosidase Crystal Type 1
Organism: Meripilus giganteus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: CL, LXE |

