Search Count: 12
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-05-22 Classification: HORMONE Ligands: ZN, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: MG, GNP, GDP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: GDP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: GDP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: GDP, TVT |
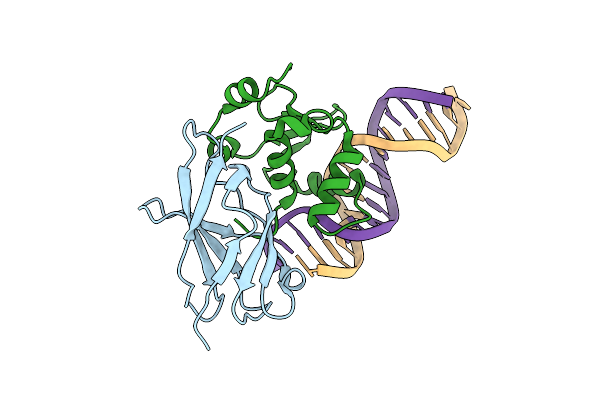 |
Structural Basis For Reactivation Of -146C>T Mutant Tert Promoter By Cooperative Binding Of P52 And Ets1/2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2018-09-19 Classification: TRANSCRIPTION/DNA |
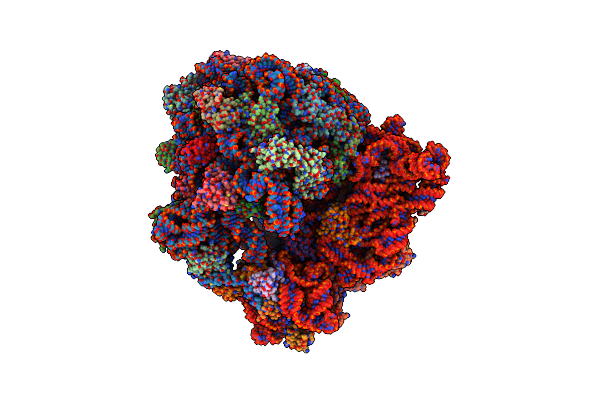 |
Structure Of A Hibernating 100S Ribosome Reveals An Inactive Conformation Of The Ribosomal Protein S1 - 70S Hibernating E. Coli Ribosome
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2018-09-05 Classification: RIBOSOME |
 |
Structure Of A Hibernating 100S Ribosome Reveals An Inactive Conformation Of The Ribosomal Protein S1 - Full 100S Hibernating E. Coli Ribosome
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2018-09-05 Classification: RIBOSOME |
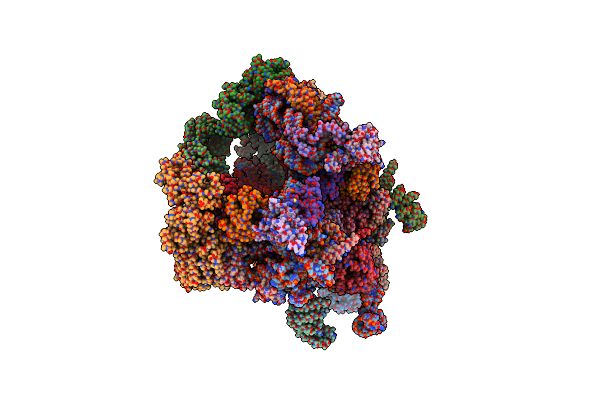 |
Organism: Chaetomium thermophilum, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Chaetomium thermophilum
Method: ELECTRON MICROSCOPY Release Date: 2016-07-27 Classification: RIBOSOME |

