Search Count: 15
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: CHAPERONE Ligands: MG |
 |
Cryo-Em Structure Of The Agr2 Dimer In Complex With The Monomeric Ire1Beta Luminal Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: CHAPERONE |
 |
Organism: Porites astreoides
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-08-26 Classification: CELL CYCLE |
 |
Homohexameric Structure Of The Second Vanadate-Dependent Bromoperoxidase (Anii) From Ascophyllum Nodosum
Organism: Ascophyllum nodosum
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2016-08-03 Classification: OXIDOREDUCTASE Ligands: VO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-11-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-02 Classification: CHAPERONE/SIGNALING PROTEIN |
 |
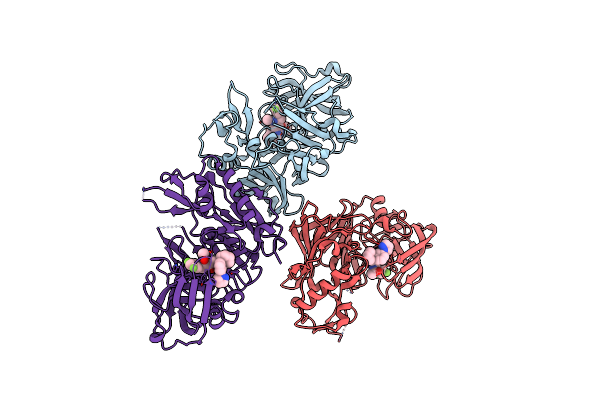
Design And Synthesis Of Hydroxyethylamine (Hea) Bace-1 Inhibitors: Prime Side Chromane-Containing Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-03-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: C6A |
 |
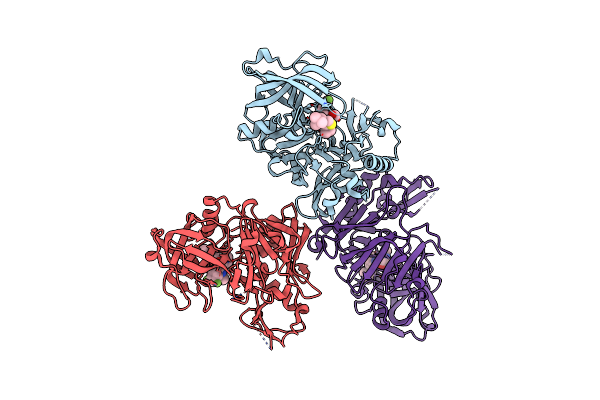
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-11-24 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: 842 |
 |
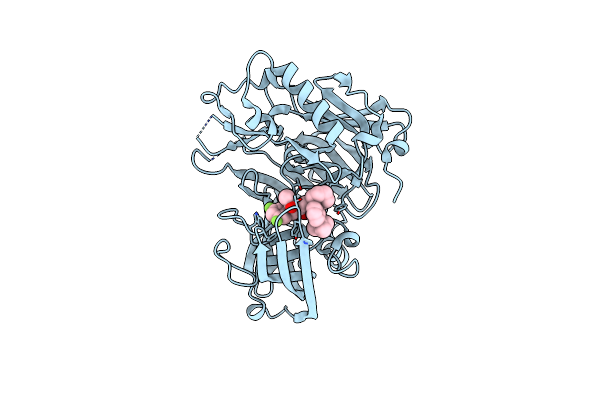
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-09-22 Classification: HYDROLASE Ligands: 957 |
 |
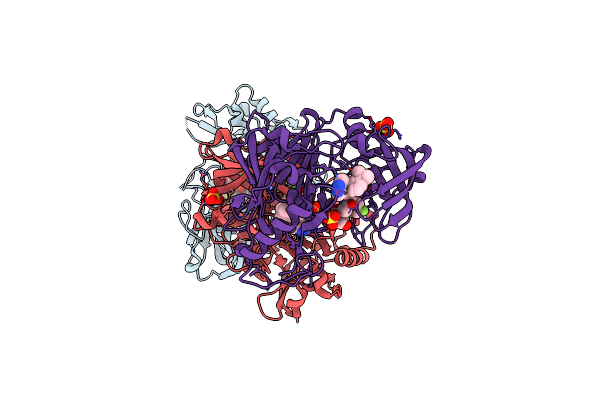
Design And Synthesis Of Potent Bace-1 Inhibitors With Cellular Activity: Structure-Activity Relationship Of P1 Substituents
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: 1LI |
 |
Design And Synthesis Of Potent Bace-1 Inhibitors With Cellular Activity: Structure-Activity Relationship Of P1 Substituents
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-01-05 Classification: HYDROLASE Ligands: SO4, GOL, 2LI |
 |
Organism: Leptospira santarosai serovar shermani
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-11-24 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
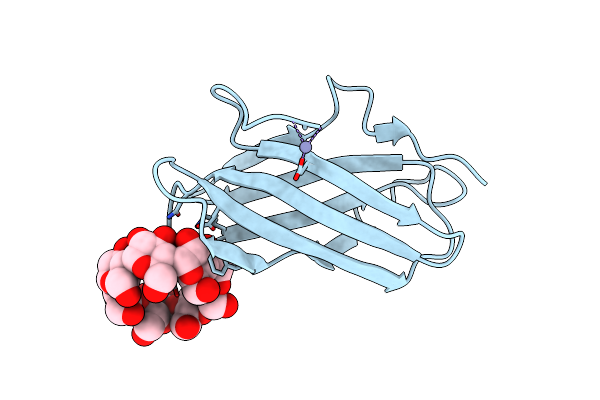
Carbohydrate-Binding Of The Starch Binding Domain Of Rhizopus Oryzae Glucoamylase In Complex With Beta-Cyclodextrin And Maltoheptaose
Organism: Rhizopus oryzae
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2009-07-07 Classification: HYDROLASE |
 |
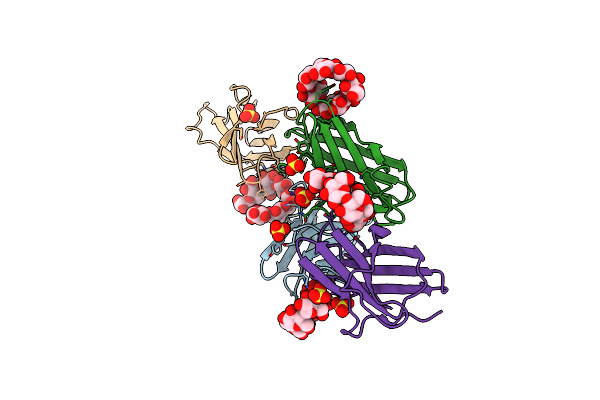
Carbohydrate-Binding Of The Starch Binding Domain Of Rhizopus Oryzae Glucoamylase In Complex With Beta-Cyclodextrin And Maltoheptaose
Organism: Rhizopus oryzae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-08-19 Classification: HYDROLASE Ligands: ZN |
 |
Carbohydrate-Binding Of The Starch Binding Domain Of Rhizopus Oryzae Glucoamylase In Complex With Beta-Cyclodextrin And Maltoheptaose
Organism: Rhizopus oryzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-08-19 Classification: HYDROLASE Ligands: SO4 |

