Search Count: 36
 |
Organism: Human herpesvirus 6b (strain hst)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-04-20 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-04-24 Classification: LIGASE Ligands: ZN, GOL, BR, CL, ACT, DTT |
 |
Crystal Structure Of E. Coli Aspartate Transcarbamoylase K164E/E239K Mutant In An Intermediate State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-05-02 Classification: TRANSFERASE/PROTEIN BINDING Ligands: ZN |
 |
Nickel Binding Domain Of Nikr From Helicobacter Pylori Disclosing Partial Metal Occupancy
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2012-04-04 Classification: METAL BINDING PROTEIN Ligands: NI, SO4 |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2011-11-23 Classification: TRANSCRIPTION Ligands: NI |
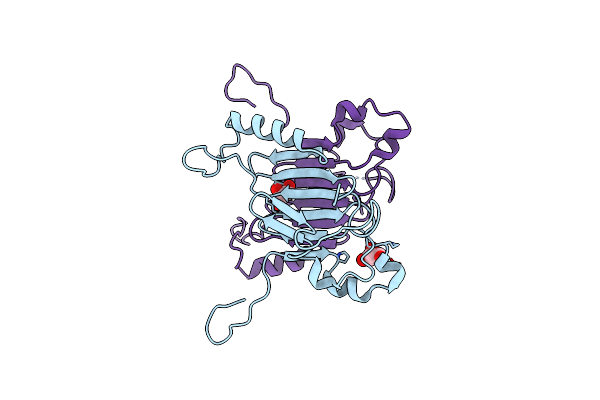 |
Crystal Structure Of The C-Terminal Domain Of Nuclear Pore Complex Component Nup116 From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-08-04 Classification: PROTEIN TRANSPORT Ligands: GOL |
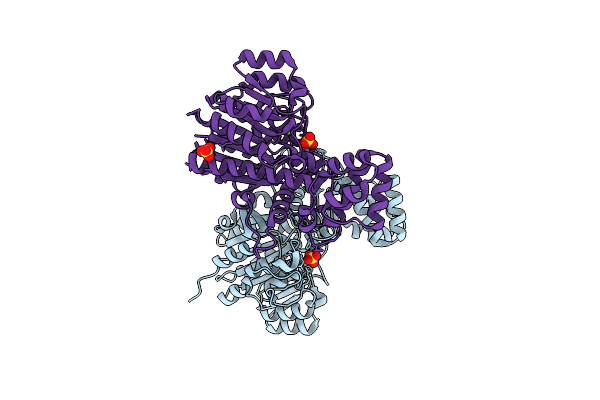 |
Crystal Structure Of Cold-Active Alkailne Phosphatase From Psychrophile Shewanella Sp.
Organism: Shewanella
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-04-14 Classification: HYDROLASE Ligands: SO4, ZN, MG |
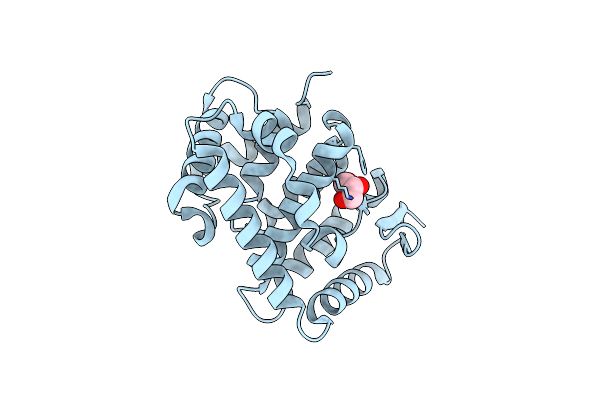 |
Crystal Structure Of The C-Terminal Domain From The Nuclear Pore Complex Component Nup133 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-26 Classification: PROTEIN TRANSPORT Ligands: GOL |
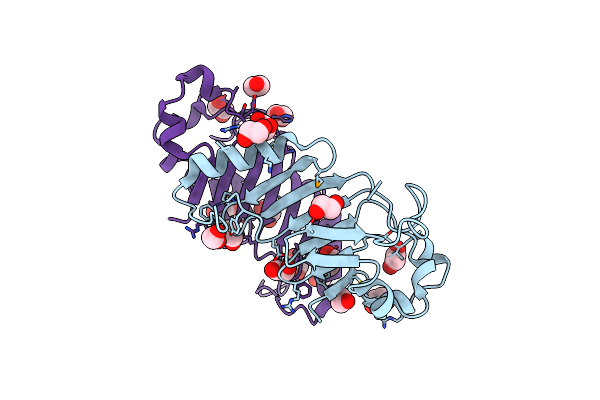 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae In The Hexagonal, P61 Space Group
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-12-23 Classification: LYASE |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-12-23 Classification: LYASE |
 |
Crystal Structure Of Acetoacetate Decarboxylase From Chromobacterium Violaceum In Complex With Acetyl Acetone Schiff Base Intermediate
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-12-23 Classification: LYASE Ligands: PNH |
 |
Structures And Conformations In Solution Of The Signal Recognition Particle Receptor From The Archaeon Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2008-11-18 Classification: TRANSPORT PROTEIN Ligands: GDP, MG |
 |
Structures And Conformations In Solution Of The Signal Recognition Particle Receptor From The Archaeon Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-11-11 Classification: TRANSPORT PROTEIN Ligands: PO4, MPD |
 |
Structures And Conformations In Solution Of The Signal Recognition Particle Receptor From The Archaeon Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2008-11-11 Classification: TRANSPORT PROTEIN Ligands: SO4, GOL |
 |
Organism: Bacteriophage hk97
Method: ELECTRON MICROSCOPY Release Date: 2008-11-04 Classification: VIRUS |
 |
Organism: Plexaura homomalla
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2008-10-07 Classification: LYASE, OXIDOREDUCTASE Ligands: FE2, HEM |
 |
Organism: Shewanella sp.
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2008-07-29 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Shewanella sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-07-29 Classification: HYDROLASE Ligands: ZN |

