Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-03-01 Classification: TOXIN Ligands: EDO |
 |
Crystal Structure Of C. Perfringens Iota-Like Enterotoxin Cpile-A With Nad+
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-01 Classification: TOXIN Ligands: NAD |
 |
Crystal Structure Of C. Perfringens Iota-Like Enterotoxin Cpile-A With Nadh
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-03-01 Classification: TOXIN Ligands: NAI |
 |
The Complex Structure Of C3Cer Exoenzyme And Gtp Bound Rhoa (Nadh-Free State)
Organism: Homo sapiens, Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-06-24 Classification: SIGNALING PROTEIN/Transferase Ligands: GSP, MG, EDO |
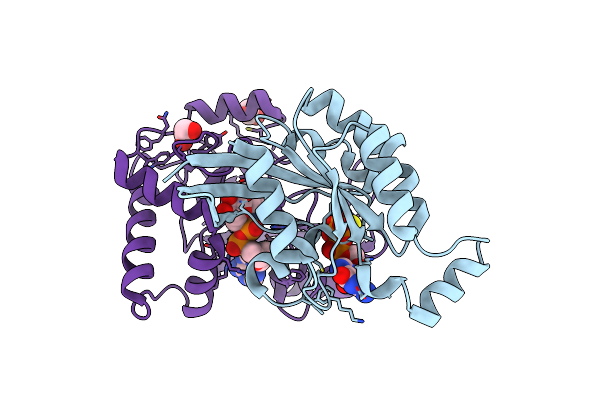 |
The Complex Structure Of C3Cer Exoenzyme And Gtp Bound Rhoa (Nadh-Bound State)
Organism: Homo sapiens, Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-06-24 Classification: SIGNALING PROTEIN/Transferase Ligands: GSP, MG, NAI, EDO |
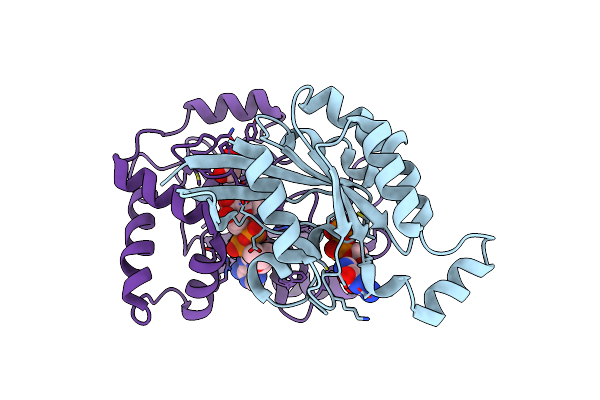 |
The Complex Structure Of C3Cer Exoenzyme And Gdp Bound Rhoa (Nadh-Bound State)
Organism: Homo sapiens, Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-06-24 Classification: SIGNALING PROTEIN/Transferase Ligands: GDP, MG, NAI, EDO |
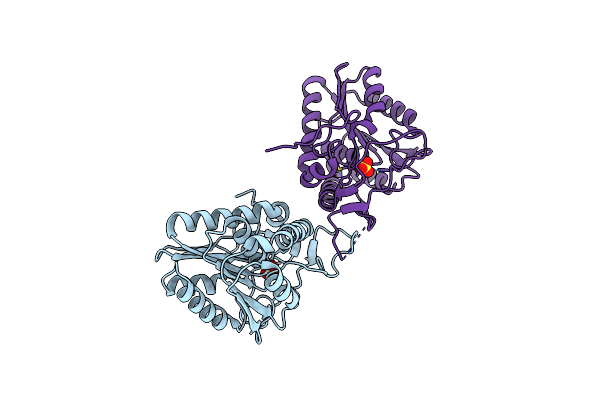 |
Organism: Bacillus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2014-07-02 Classification: HYDROLASE Ligands: CL, SO4 |
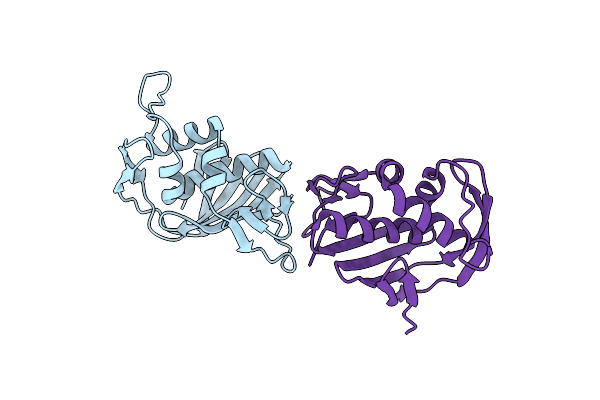 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2014-02-05 Classification: VIRAL PROTEIN |
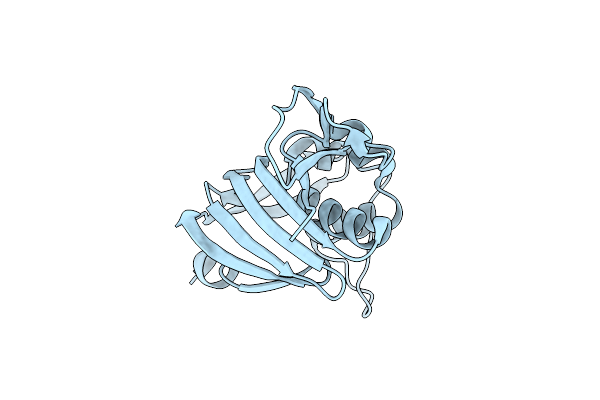 |
P453H/I471T Mutant Of Pb2 Middle Domain From Influenza Virus A/Puerto Rico/8/34(H1N1)
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-12-18 Classification: VIRAL PROTEIN |
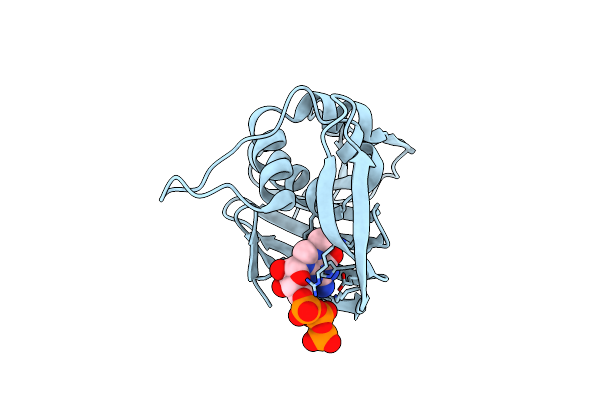 |
P453H/I471T Mutant Of Pb2 Middle Domain From Influenza Virus A/Puerto Rico/8/34(H1N1) With M7Gtp
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2013-12-18 Classification: VIRAL PROTEIN Ligands: MGT |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-02-20 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: PO4, CA, ATP, LAR |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-02-20 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: NAD, PO4, EDO, CA, ATP, LAR |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-02-20 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: PO4, EDO, AR6, CA, LAR, ATP |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2013-02-20 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: NAD, PO4, EDO, CA, ATP, LAR |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2013-02-20 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: NAD, PO4, EDO, CA, ATP, LAR |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2013-02-20 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: NAD, PO4, EDO, CA, ATP, LAR |
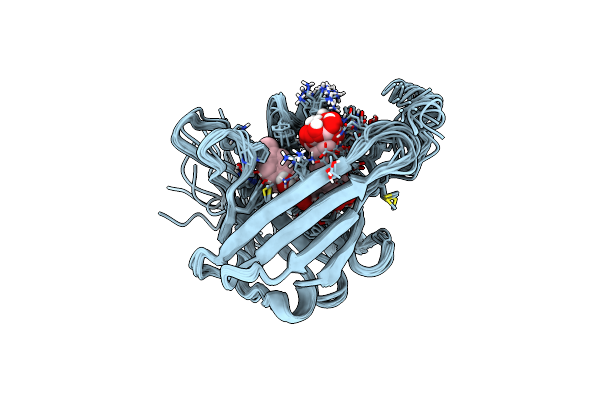 |
Solution Structure Of Mouse Lipocalin-Type Prostaglandin D Synthase / Substrate Analog (U-46619) Complex
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2011-02-02 Classification: ISOMERASE Ligands: PUC |
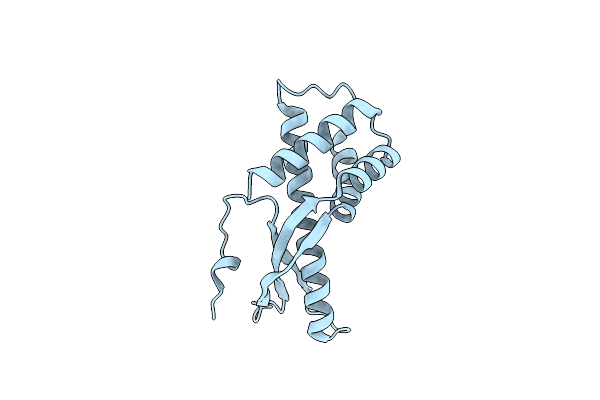 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2009-09-22 Classification: TOXIN |
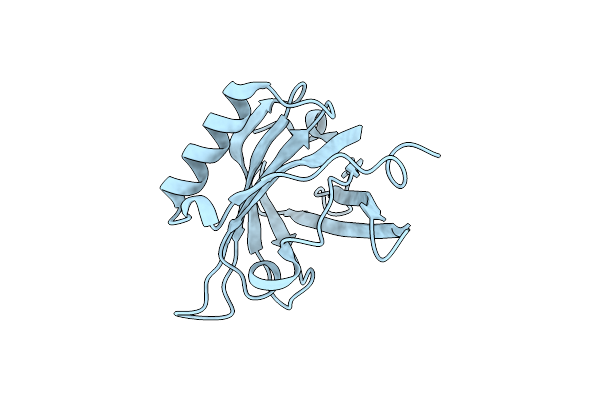 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-10-03 Classification: ISOMERASE |
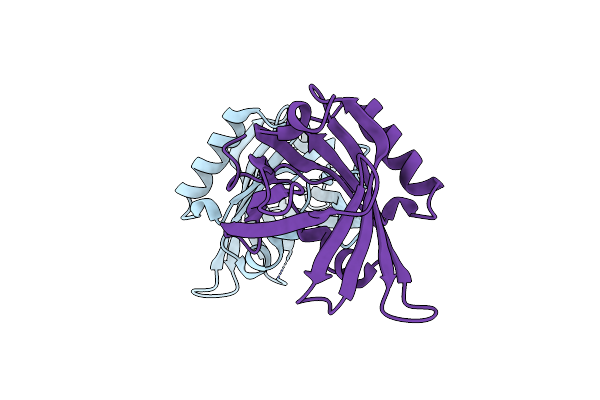 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-10-03 Classification: ISOMERASE |