Search Count: 14
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
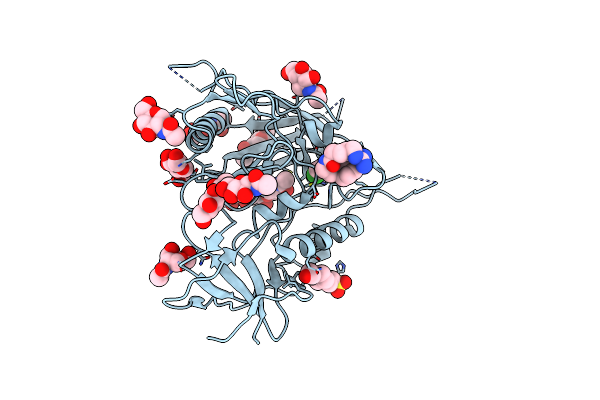
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-12-13 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
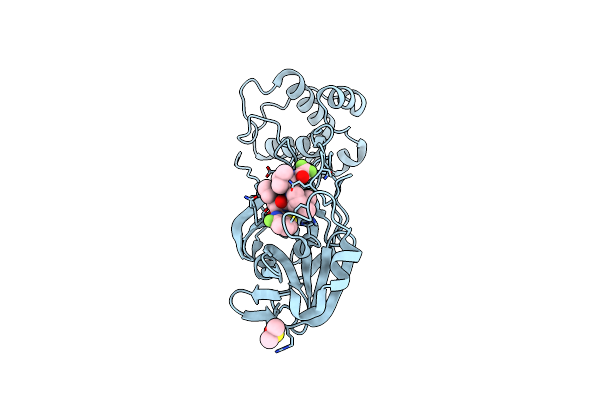
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Yir-821
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-01-11 Classification: VIRAL PROTEIN Ligands: NAG, R5K, EPE |
 |
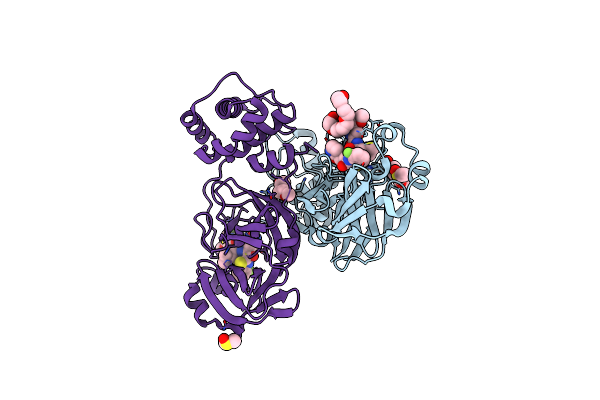
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-09-21 Classification: HYDROLASE/INHIBITOR Ligands: T2L, DMS |
 |
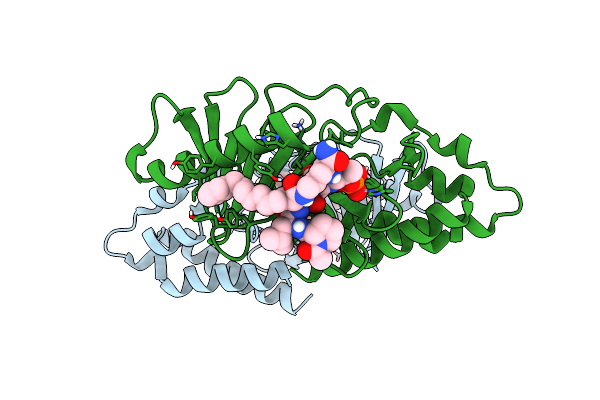
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-198
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-08-24 Classification: HYDROLASE/INHIBITOR Ligands: 1PE, MES, DMS, PG0, T1X |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Inhibitor Tkb-248
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-08-24 Classification: HYDROLASE/INHIBITOR Ligands: T43, DMS, ZN |
 |
Plk-1 Polo-Box Domain In Complex With A High Affinity Macrocycle Synthesized Using A Novel Glutamic Acid Analog
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-03-30 Classification: PROTEIN BINDING Ligands: ZOY |
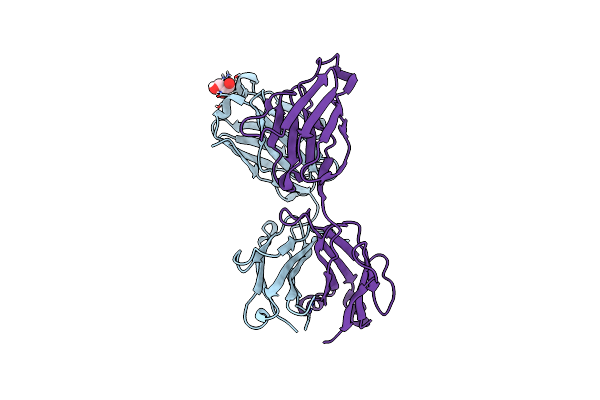 |
Site-Specific Lysine Arylation As An Alternative Bioconjugation Strategy For Chemically Programmed Antibodies And Antibody-Drug Conjugates
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2019-11-13 Classification: IMMUNE SYSTEM Ligands: GOL |
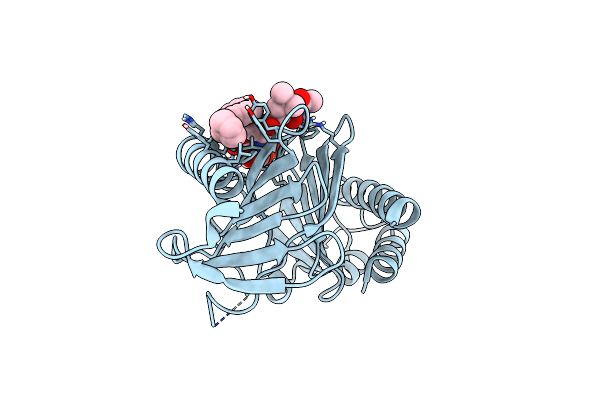 |
Plk-1 Polo-Box Domain In Complex With Histidine N(Tau)-Cyclized Macrocycle 5B.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-09-12 Classification: PROTEIN BINDING Ligands: AML |
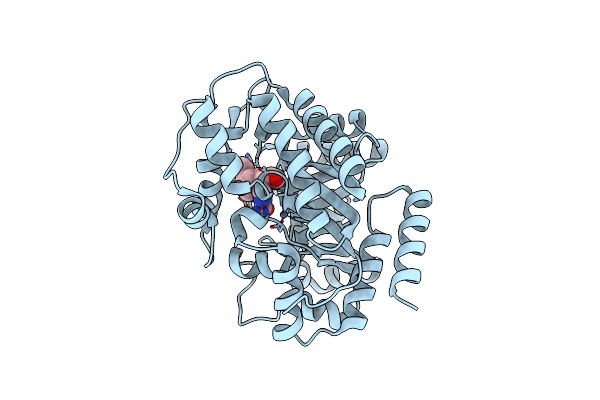 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-11-07 Classification: HYDROLASE Ligands: ZN, FR6 |
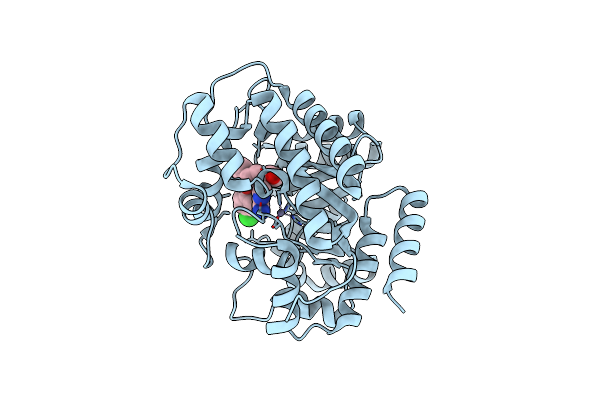 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-08-16 Classification: HYDROLASE Ligands: ZN, FRL |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-12-21 Classification: HYDROLASE Ligands: ZN, FR7 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-12-21 Classification: HYDROLASE Ligands: ZN, FRC |

