Search Count: 92
 |
Organism: Olindias formosus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN |
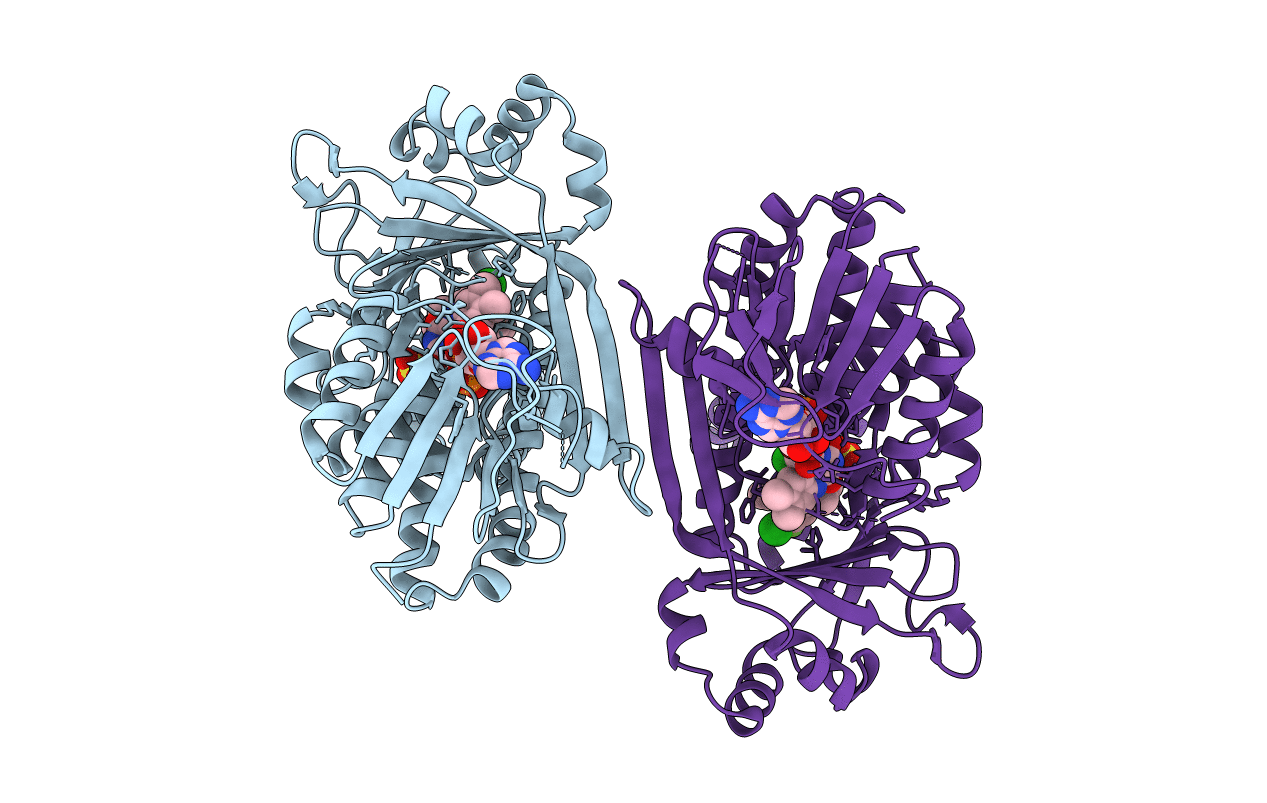 |
In Meso Full-Length Rat Kmo In Complex With A Pyrazoyl Benzoic Acid Inhibitor
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-23 Classification: MEMBRANE PROTEIN Ligands: FAD, EGO, SO4, CL |
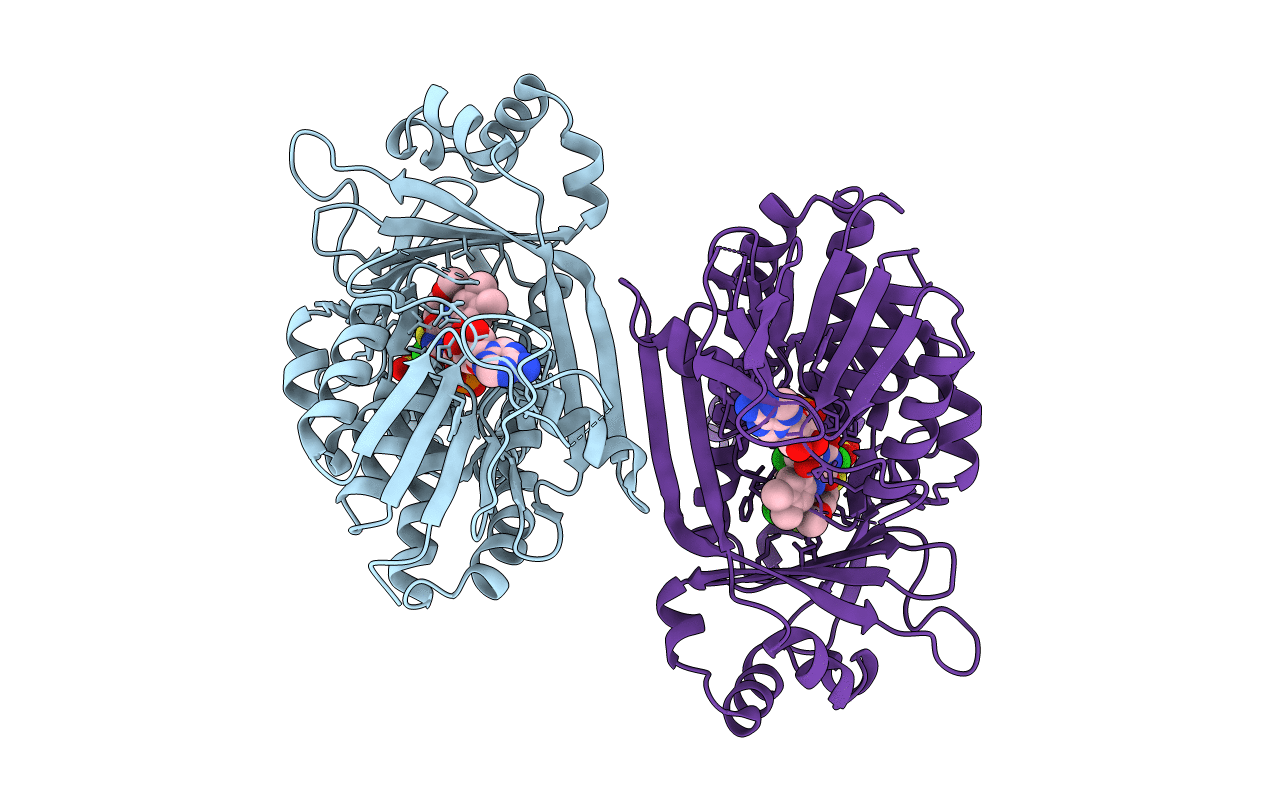 |
In Meso Full-Length Rat Kmo In Complex With An Inhibitor Identified Via Dna-Encoded Chemical Library Screening
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-23 Classification: MEMBRANE PROTEIN Ligands: FAD, SO4, CL, EGU |
 |
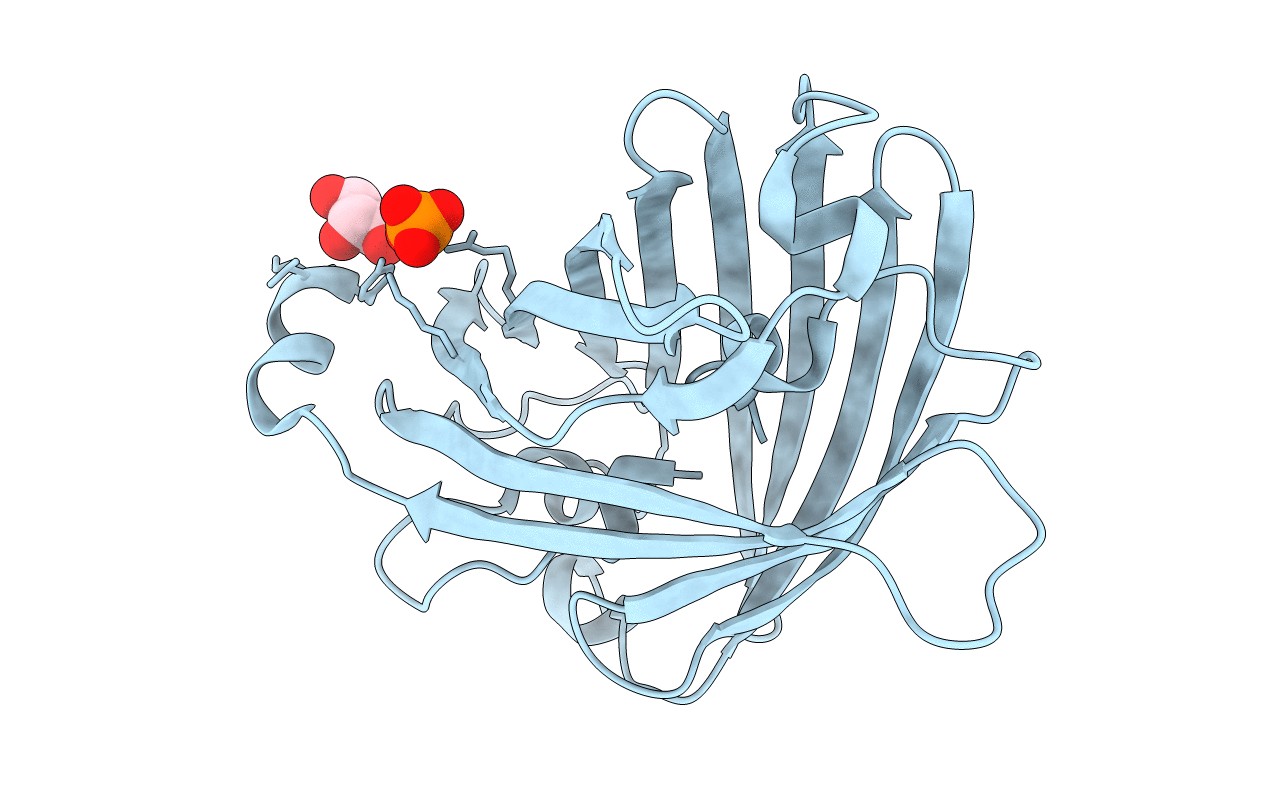
Organism: Olindias
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2019-11-06 Classification: FLUORESCENT PROTEIN Ligands: CL, GOL, PO4 |
 |
Photoswitchable Fluorescent Protein Gamillus, N150C/T204V Double Mutant, On-State
Organism: Olindias
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-11-06 Classification: FLUORESCENT PROTEIN Ligands: PO4, GOL, CL |
 |
Photoswitchable Fluorescent Protein Gamillus, N150C/T204V Double Mutant, Off-State
Organism: Olindias
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2019-11-06 Classification: FLUORESCENT PROTEIN Ligands: CL, GOL |
 |
Organism: Olindias
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-06 Classification: FLUORESCENT PROTEIN Ligands: PO4, GOL, CL |
 |
Structure Of Acetophenone Reductase From Geotrichum Candidum Nbrc 4597 In Complex With Nad
Organism: Geotrichum candidum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-09-11 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
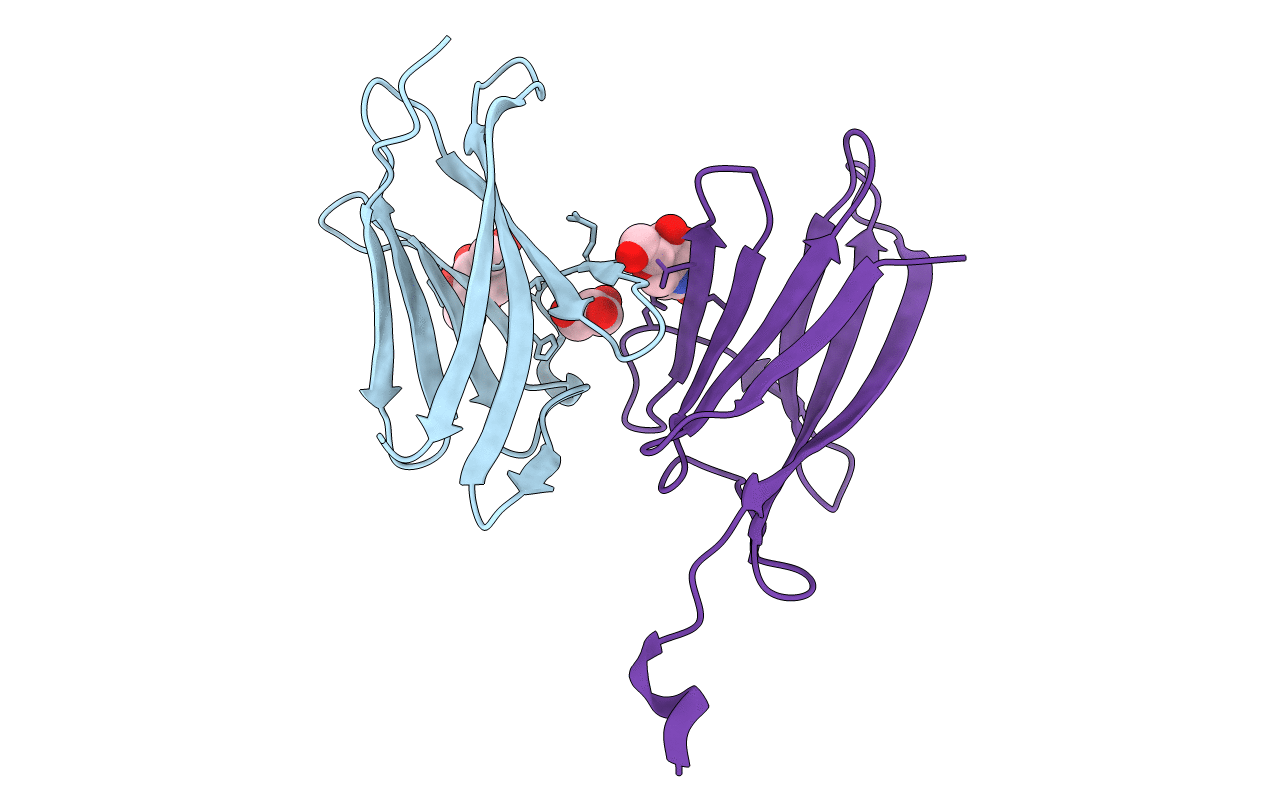 |
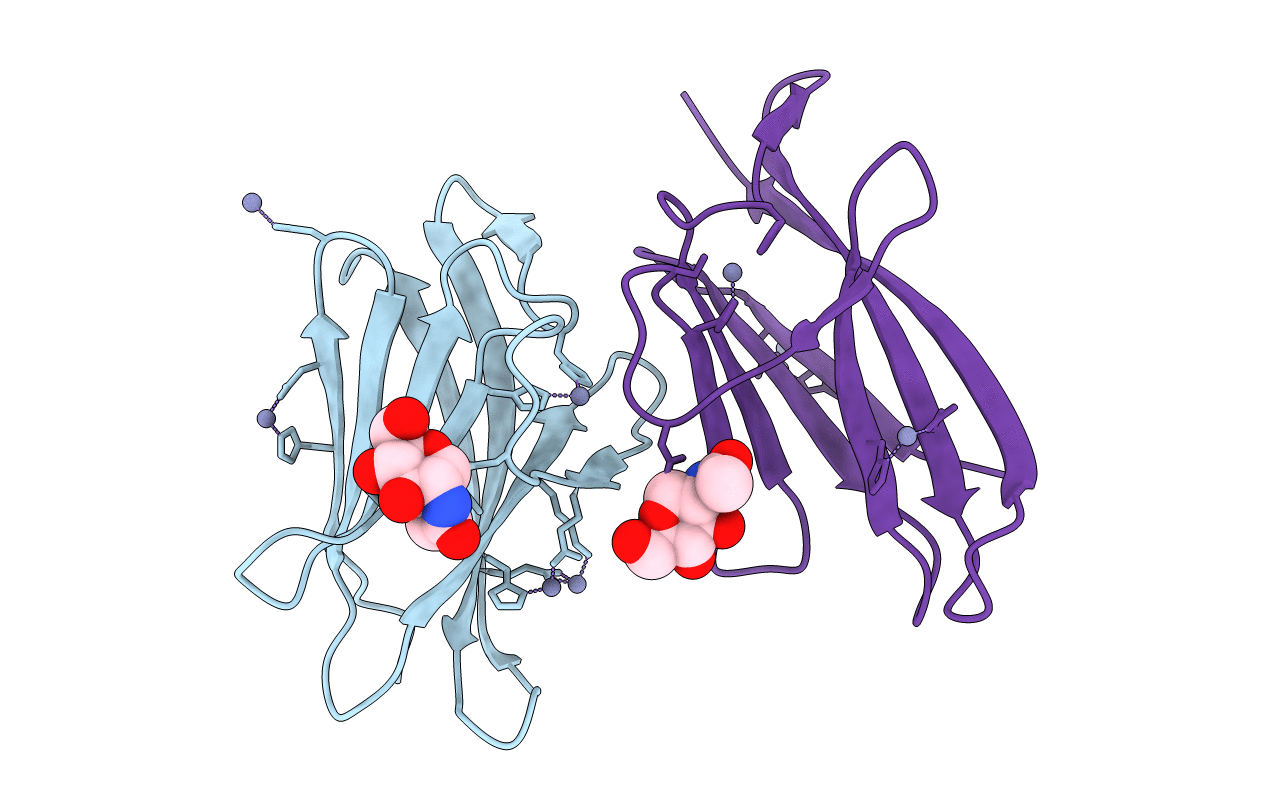
Molecular Basis Of Egg Coat Filament Cross-Linking: High-Resolution Structure Of The Partially Deglycosylated Zp1 Zp-N1 Domain Homodimer
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-19 Classification: CELL ADHESION Ligands: NAG, GOL |
 |
Molecular Basis Of Egg Coat Filament Cross-Linking: Zn-Sad Structure Of The Partially Deglycosylated Zp1 Zp-N1 Domain Homodimer
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-06-19 Classification: CELL ADHESION Ligands: NAG, ZN |
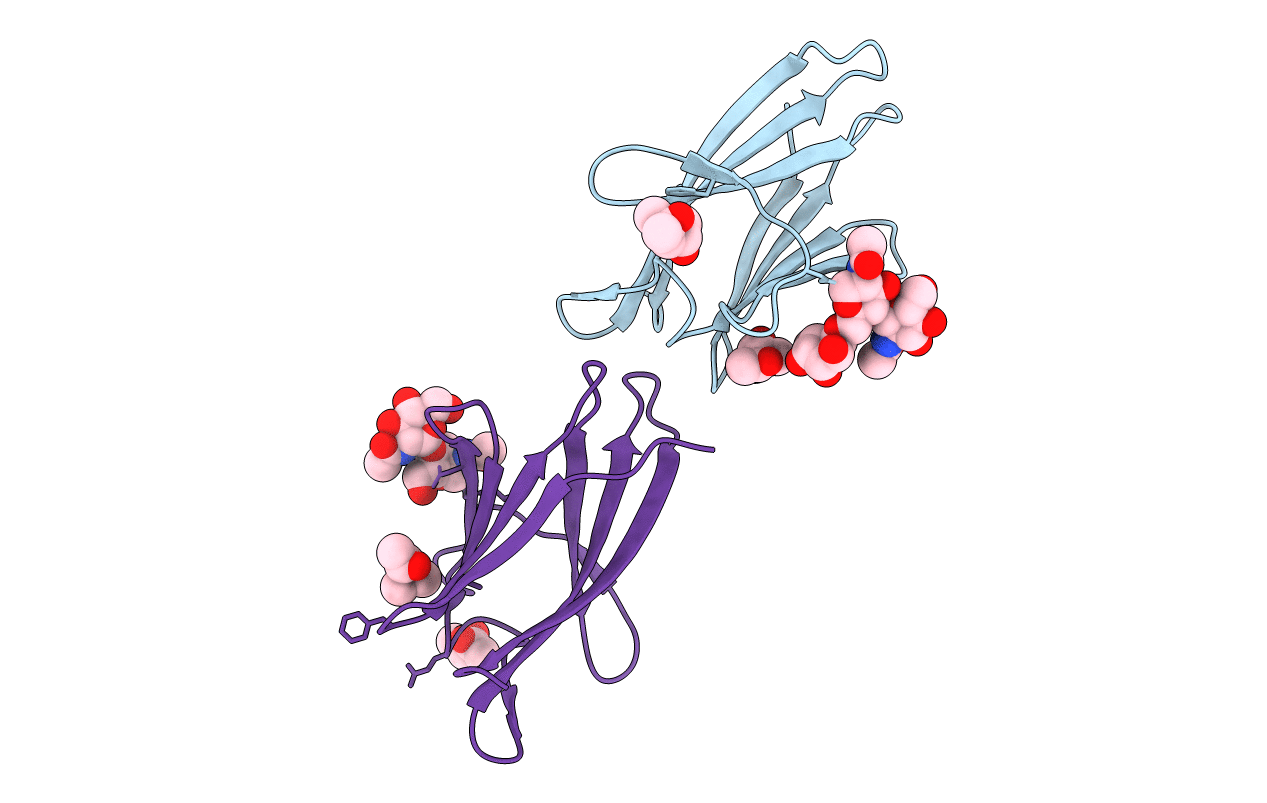 |
Molecular Basis Of Egg Coat Filament Cross-Linking: Structure Of The Glycosylated Zp1 Zp-N1 Domain Homodimer
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-06-19 Classification: CELL ADHESION Ligands: MPD |
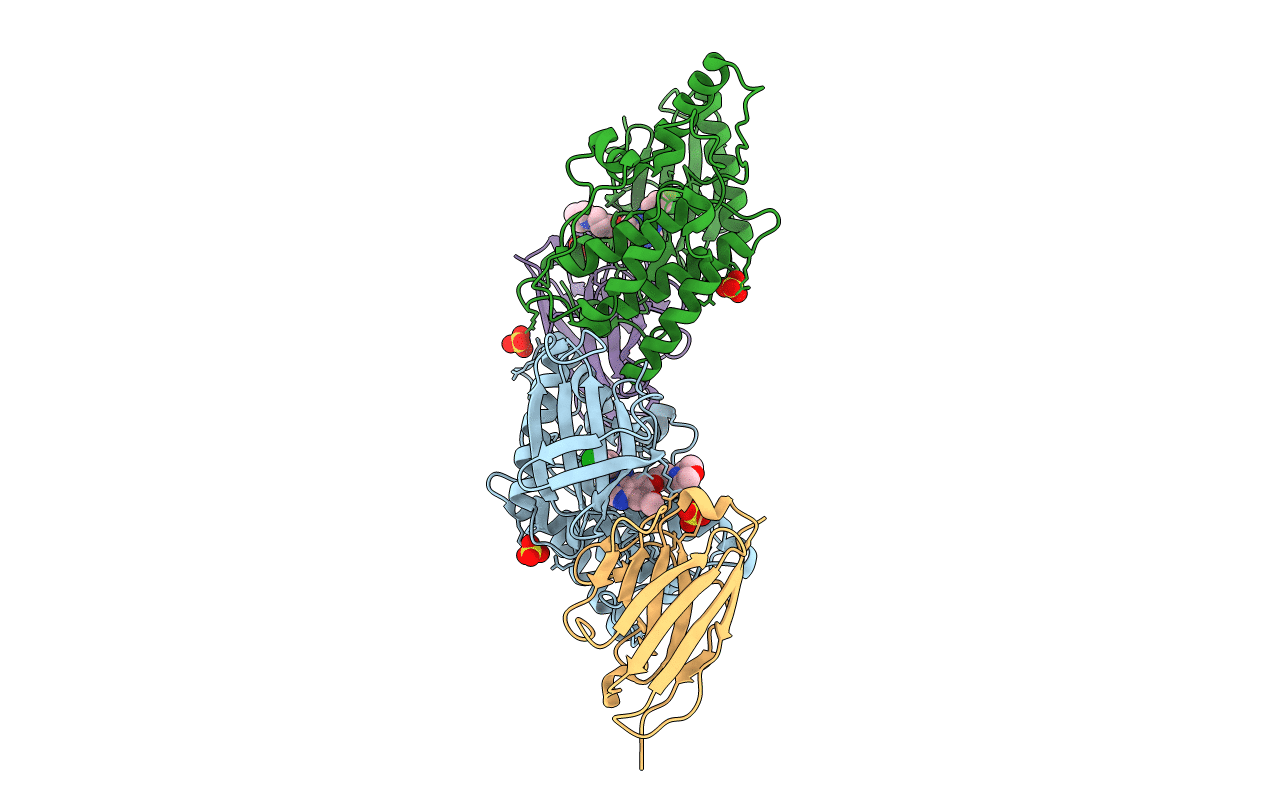 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-08-29 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: IRE, SO4 |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-08-29 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: IRE |
 |
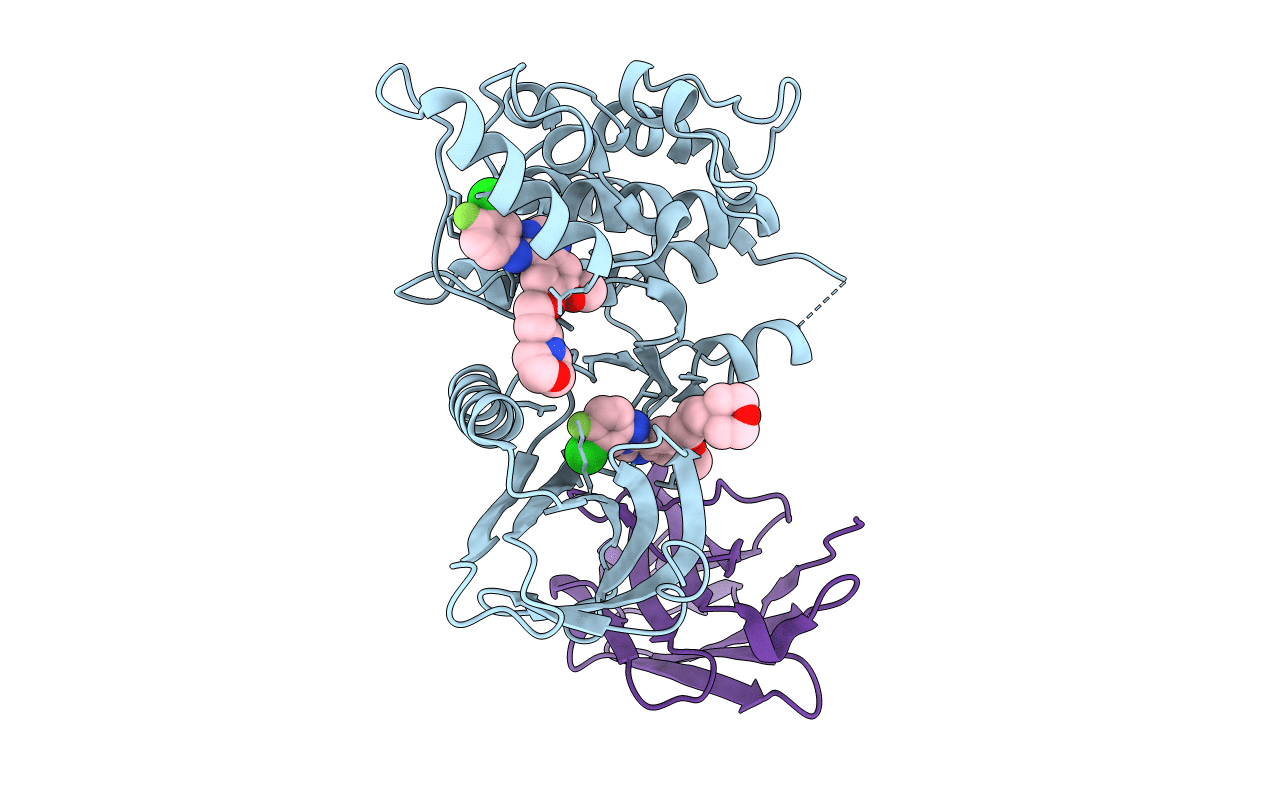
Crystal Structure Of The Complex Of Di-Acetylated Histone H4 And 2A7D9 Fab Fragment
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-22 Classification: NUCLEAR PROTEIN |
 |
Crystal Structure Of The Complex Of Di-Acetylated Histone H4 And 1A9D7 Fab Fragment
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-08-22 Classification: NUCLEAR PROTEIN Ligands: ZN |
 |
Crystal Structure Of Hck Kinase Complexed With A Pyrrolo-Pyrimidine Inhibitor 7-[Trans-4-(4-Methylpiperazin-1-Yl)Cyclohexyl]-5-(4-Phenoxyphenyl)-7H-Pyrrolo[2,3-D]Pyrimidin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: VSE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2018-02-28 Classification: SIGNALING PROTEIN Ligands: NAG, ZN |
 |
Organism: Olindias
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-01-17 Classification: FLUORESCENT PROTEIN Ligands: PO4, GOL, CL |
 |
Organism: Olindias
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2018-01-17 Classification: FLUORESCENT PROTEIN Ligands: PO4 |
 |
Organism: Listeria innocua clip11262
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-12-02 Classification: TRANSFERASE Ligands: SO4, GOL, MES |

