Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
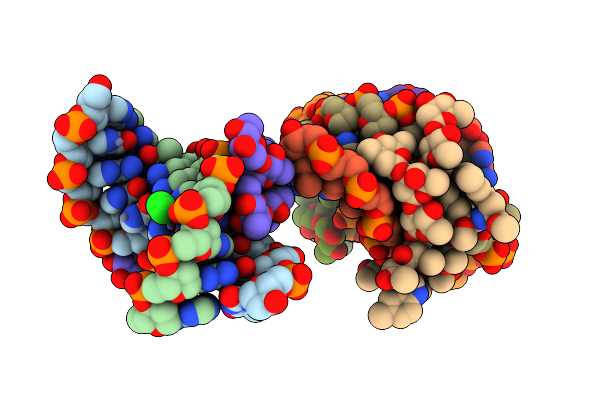 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-5-Methyl-Trp In Complex With Dna Agaggcctct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA Ligands: ZN, A1BNI, CL, NA |
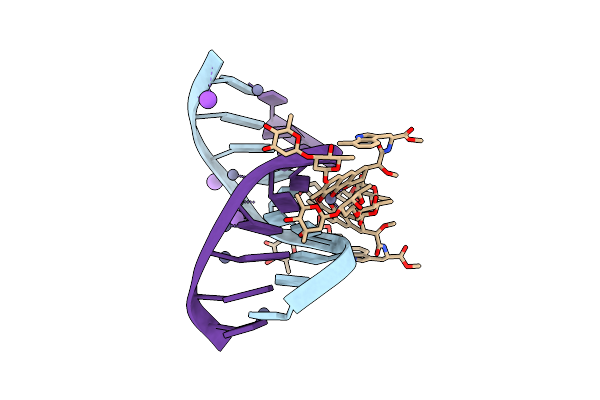 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-7-Methyl-Trp In Complex With Double-Stranded Dna Agaggcctct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA Ligands: A1BNK, ZN, NA |
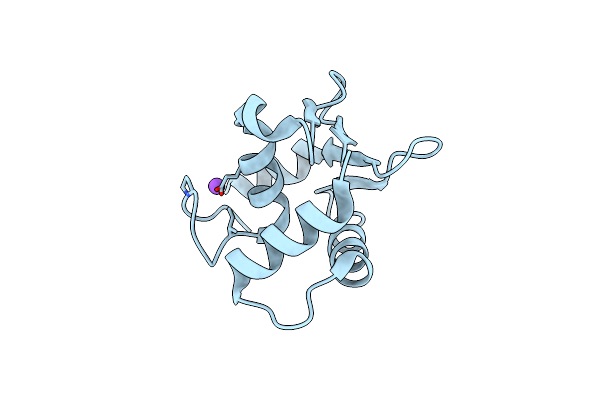 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN |
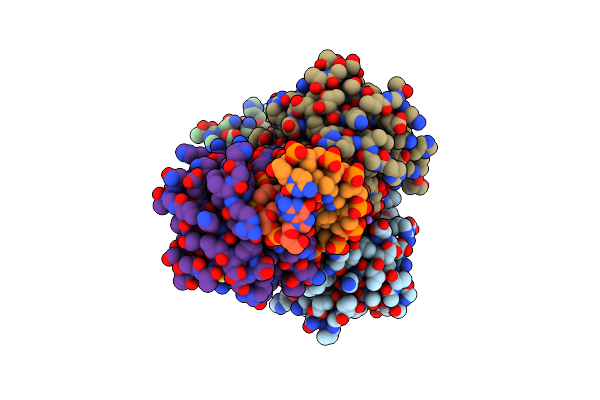 |
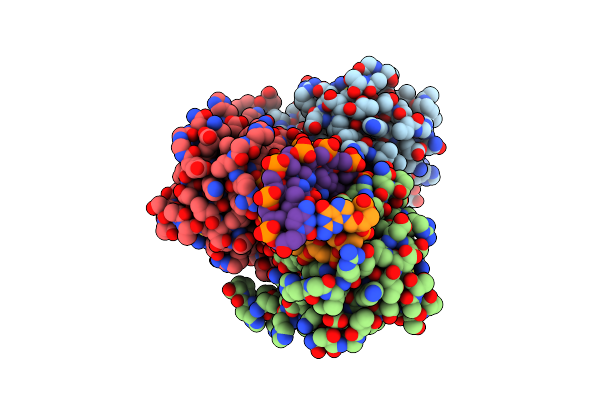
Crystal Structure Of The Dna Binding Domain Of Fli1 In Complex With A Dna Containing Four Contiguous Ggaa Sites
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN/DNA |
 |
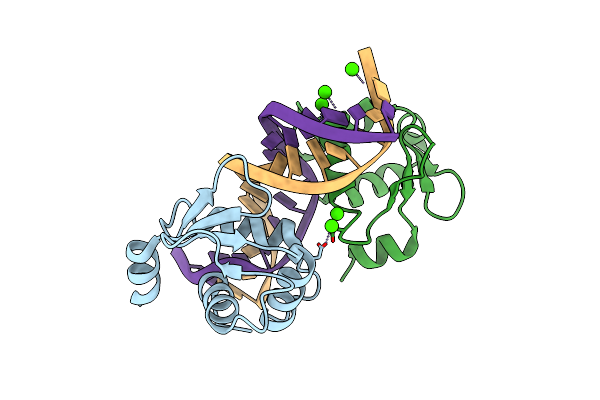
Crystal Structure Of The Dna Binding Domain Of Fli1 In Complex With A Dna Containing Three Contiguous Ggaa Sites
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN/DNA |
 |
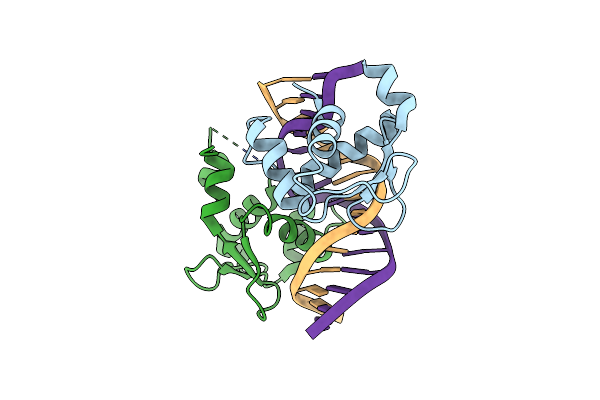
Crystal Structure Of The Dna Binding Domain Of Fli1 (F362A) In Complex With A Dna Containing Two Contiguous Ggaa Sites
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN/DNA Ligands: CA |
 |
Crystal Structure Of The Dna Binding Domain Of Fli1 (Wild-Type) In Complex With A Dna Containing Two Contiguous Ggaa Sites
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-08-07 Classification: DNA Ligands: ZN, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-08-07 Classification: DNA Ligands: ZN, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-08-07 Classification: DNA Ligands: ZN, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-07 Classification: DNA Ligands: ZN, ACT |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4, BOG |
 |
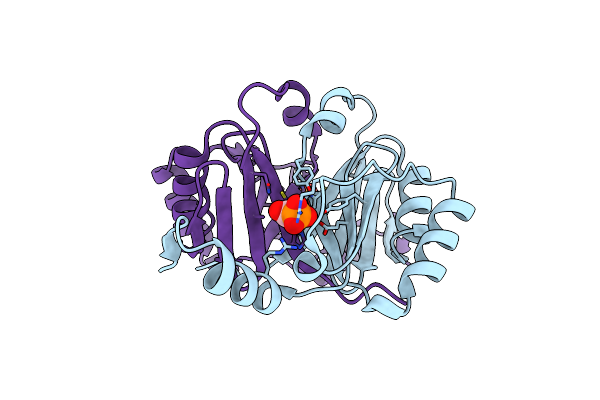
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Fosb From Bacillus Cereus With Zinc And Phosphonoformate
Organism: Bacillus cereus atcc 10987
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: PPF, ZN |
 |
Crystal Structure Of Fosb From Bacillus Cereus With Zinc And 2-Phosphonopropionic Acid
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: UXR, FMT, ZN, MG |
 |
Crystal Structure Of Fosb From Bacillus Cereus With Zinc And Phosphonoacetate
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: PAE, FMT, ZN, MG |
 |
Crystal Structure Of Fosb From Bacillus Cereus With Zinc And 1-Hydroxypropylphosphonic Acid
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: 1JJ, FMT, GOL, ZN, MG |
 |
Crystal Structure Of Fosb From Bacillus Cereus With Zinc And (1-Hydroxy-2-Methylpropyl)Phosphonic Acid
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: FMT, YS8, ZN, MG |