Search Count: 24
 |
Organism: Bos taurus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN |
 |
Cryo-Em Structure Of Rhodopsin-Gi Bound With Antibody Fragments Scfv16 And Fab79, Conformation 1
Organism: Bos taurus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN |
 |
Cryo-Em Structure Of Rhodopsin-Gi Bound With Antibody Fragments Scfv16 And Fab79, Conformation 2
Organism: Bos taurus, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Hasarius adansoni, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
 |
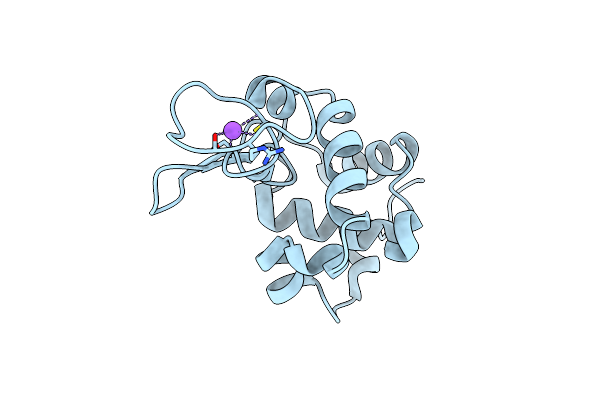
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Swissfel)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
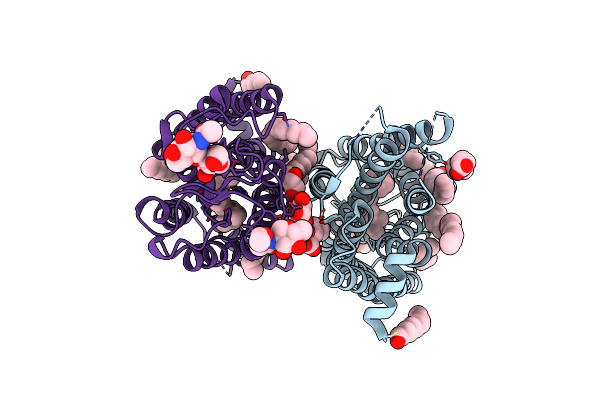 |
1 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
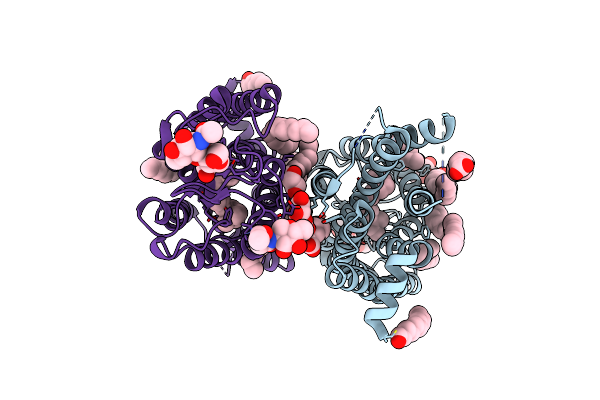 |
10 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
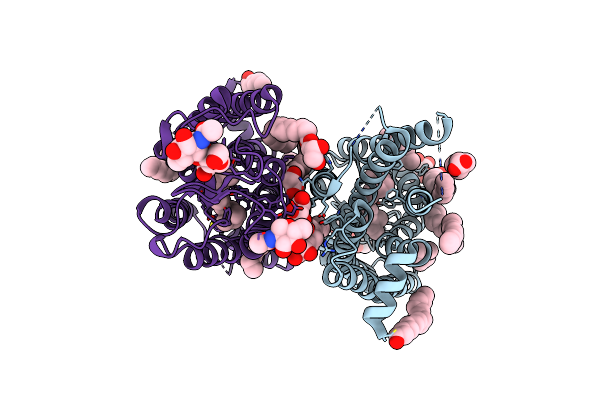 |
100 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: RET, NAG, DAO, OLC, PLM |
 |
Structure Of Hen Egg White Lysozyme Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: CL, NA |
 |
Structure Of Thaumatin Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-09-01 Classification: PLANT PROTEIN Ligands: TLA, PGO, NA |
 |
Structure Of Insulin Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2021-09-01 Classification: HORMONE Ligands: PGR, NA |
 |
Structure Of Tubulin Darpin Complex 1 Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2021-09-01 Classification: CELL CYCLE Ligands: GTP, MG, GDP, PG0 |
 |
Structure Of Sponge-Phase Grown Peptst2 Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-09-01 Classification: TRANSPORT PROTEIN Ligands: PO4, 78M, PG0, PGE |
 |
Structure Of Ribonucleotide Reductase R2 From Escherichia Coli Collected By Still Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-01 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Structure Of Ribonucleotide Reductase R2 From Escherichia Coli Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-09-01 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Structural Basis Of The Activation Of The Cc Chemokine Receptor 5 By A Chemokine Agonist
Organism: Homo sapiens, Mus musculus, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: SIGNALING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-07-10 Classification: IMMUNE SYSTEM Ligands: PG4, EDO, PGE, MLT |

