Search Count: 29
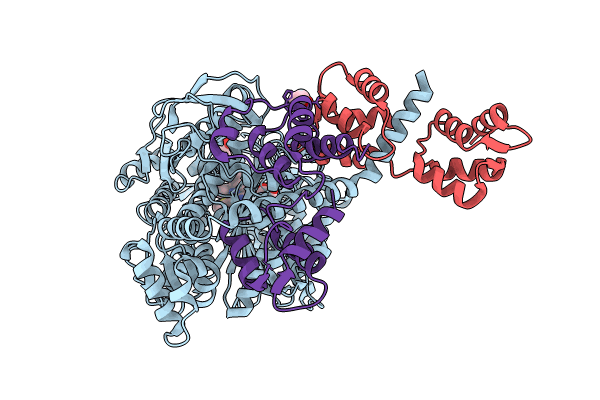 |
Plasmodium Falciparum Myosin A Full-Length, Post-Rigor State Complexed To The Inhibitor Knx-002
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-02-21 Classification: MOTOR PROTEIN Ligands: EDO, KQ0, PG4 |
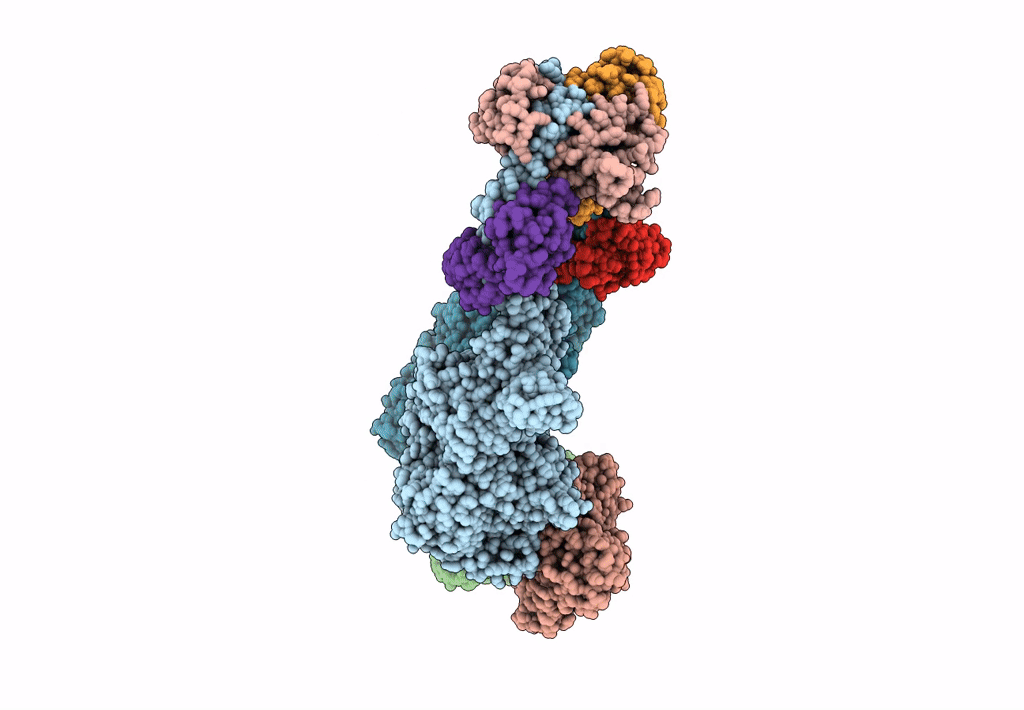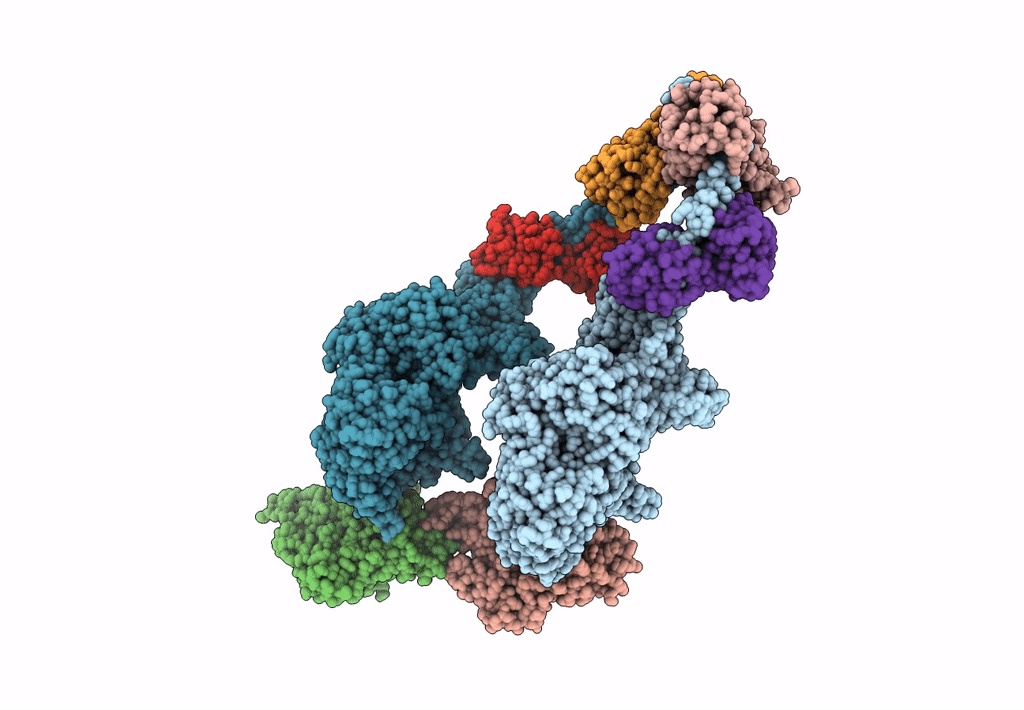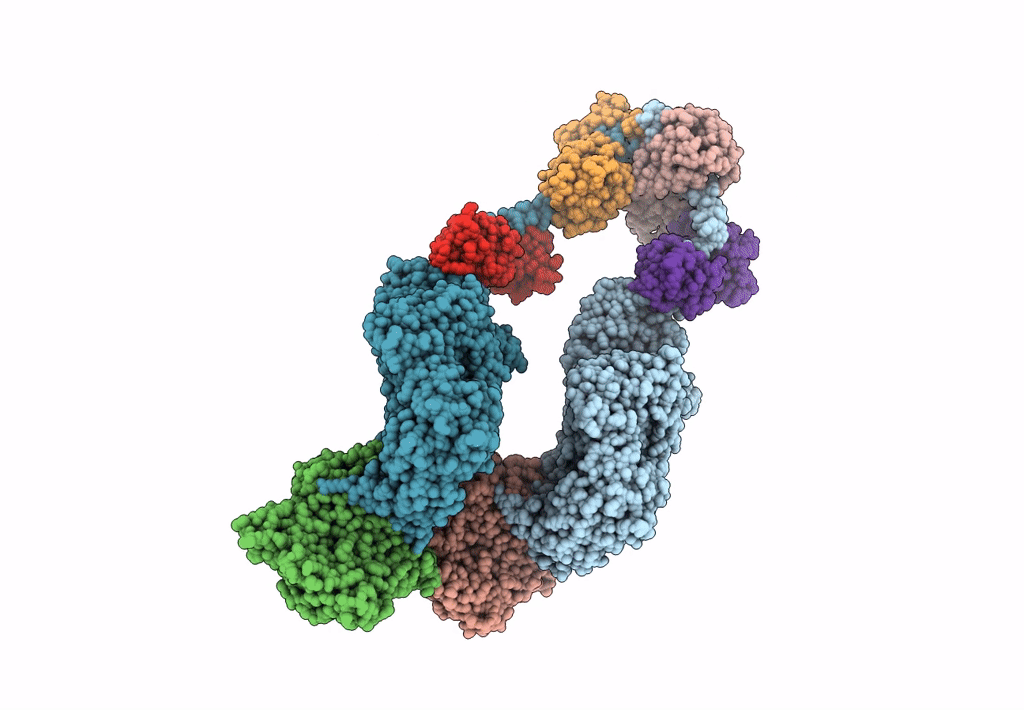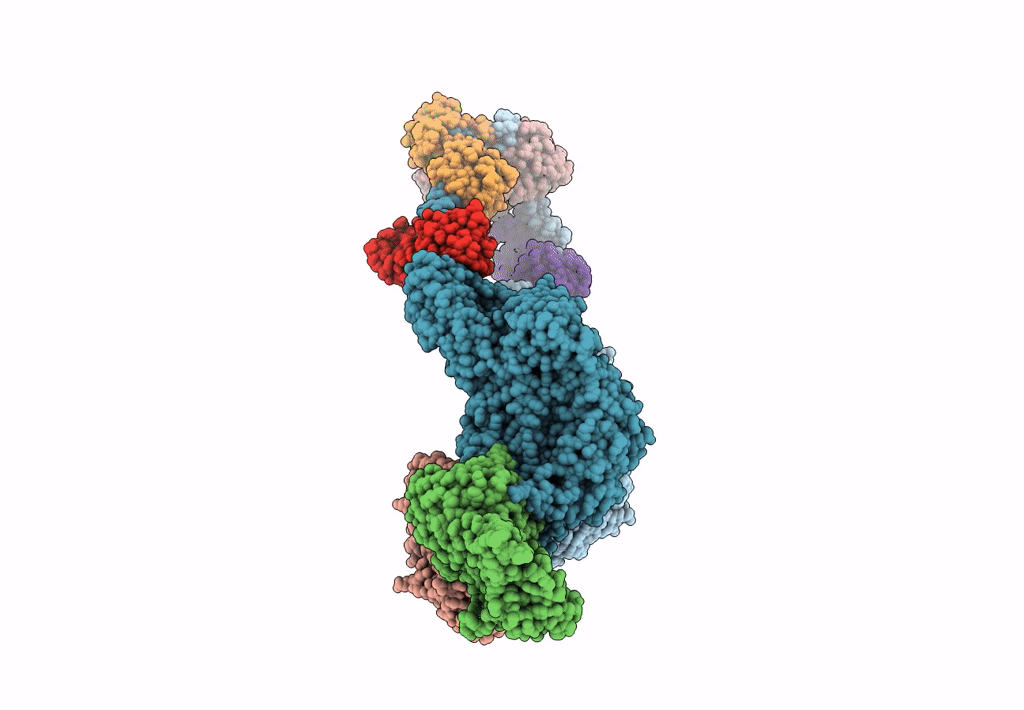 |
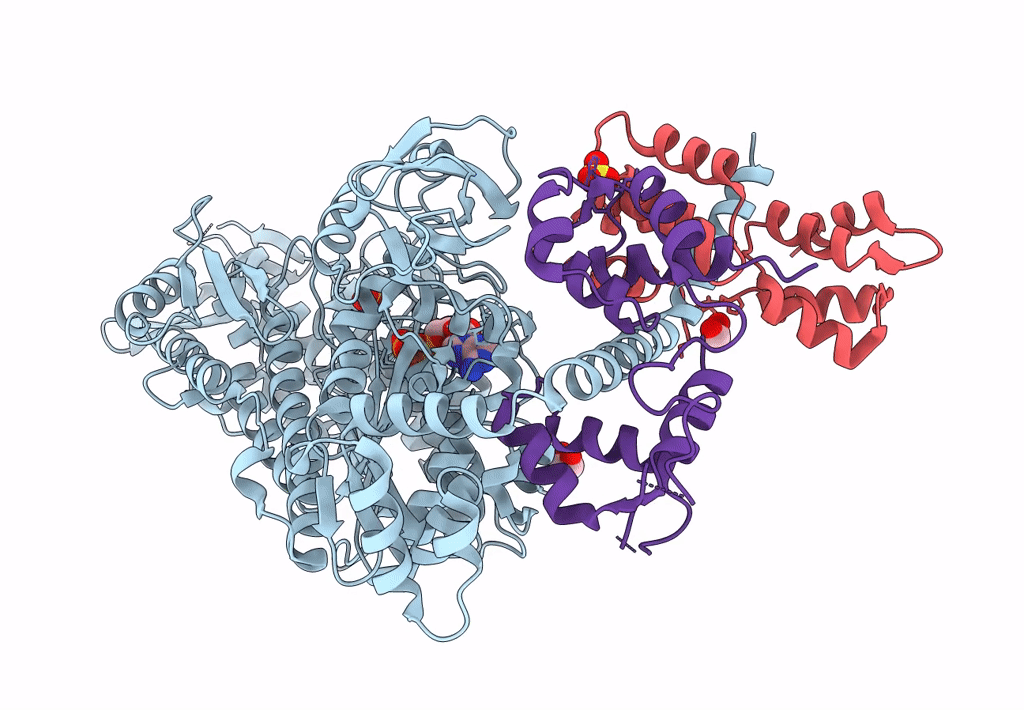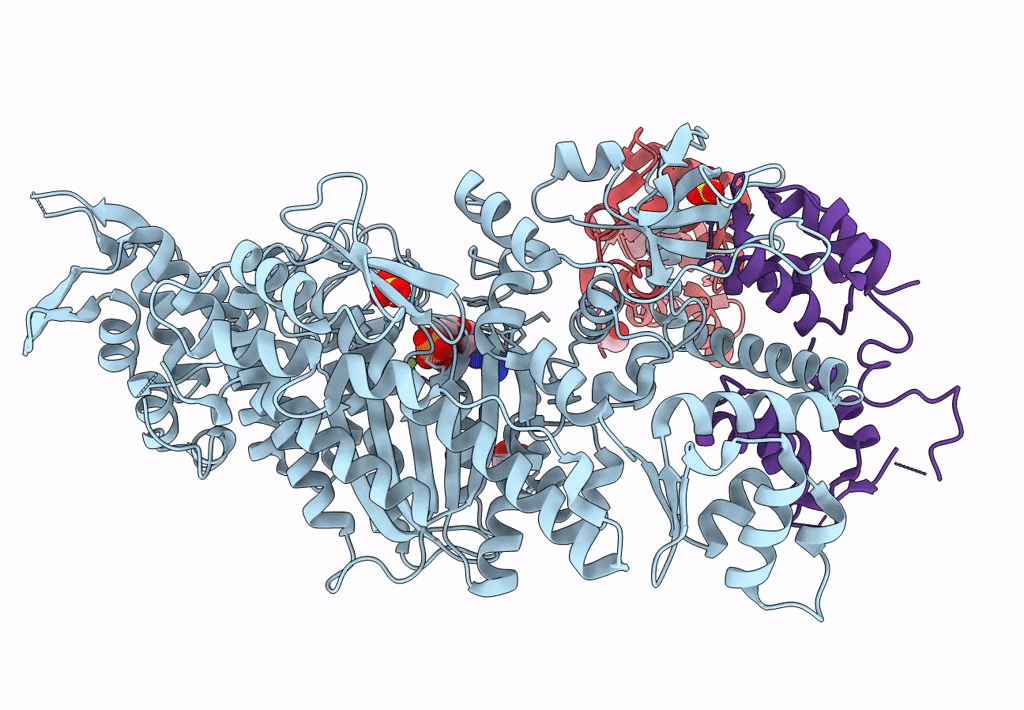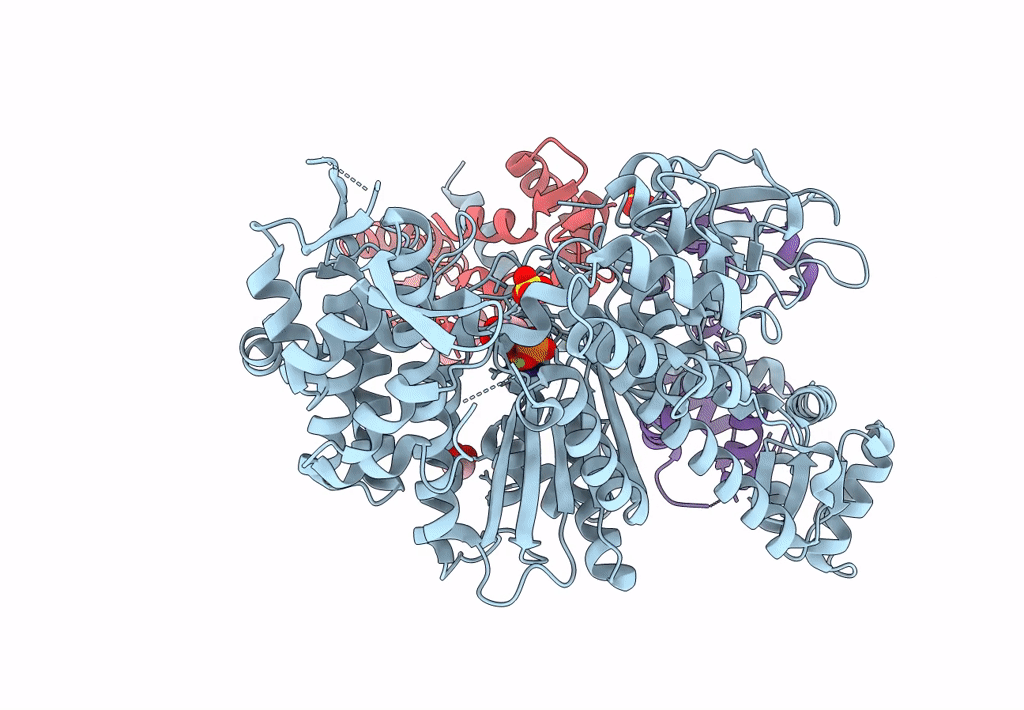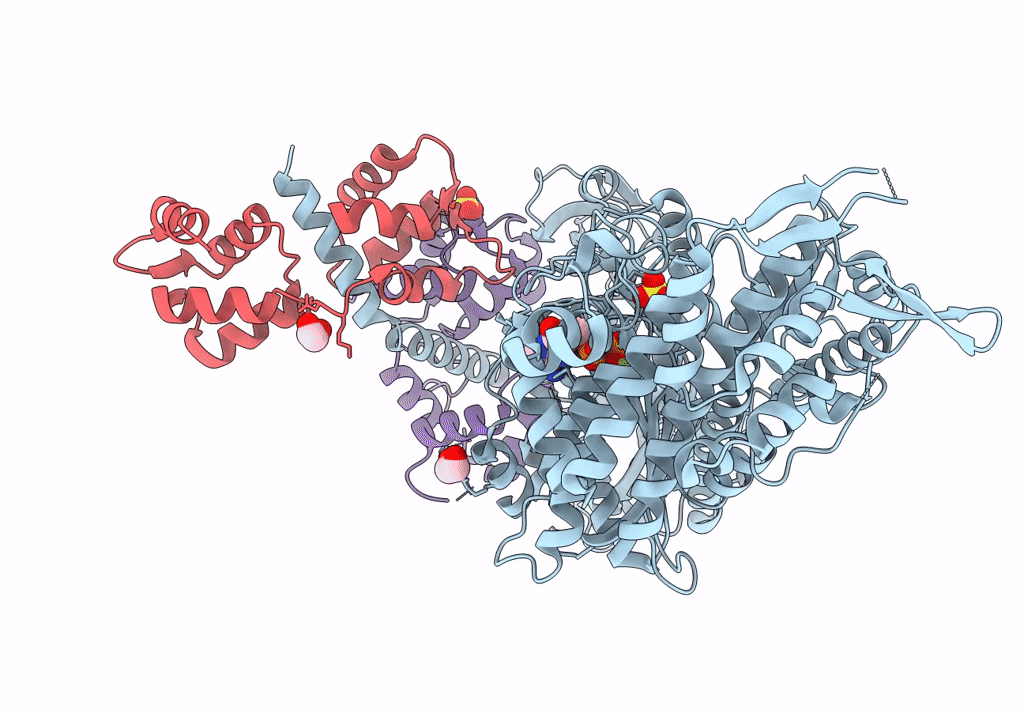
Homology Model Of Acto-Hmm Complex In Adp-State. Chicken Smooth Muscle Hmm And Chicken Pectoralis Actin
Organism: Gallus gallus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MOTOR PROTEIN |
 |
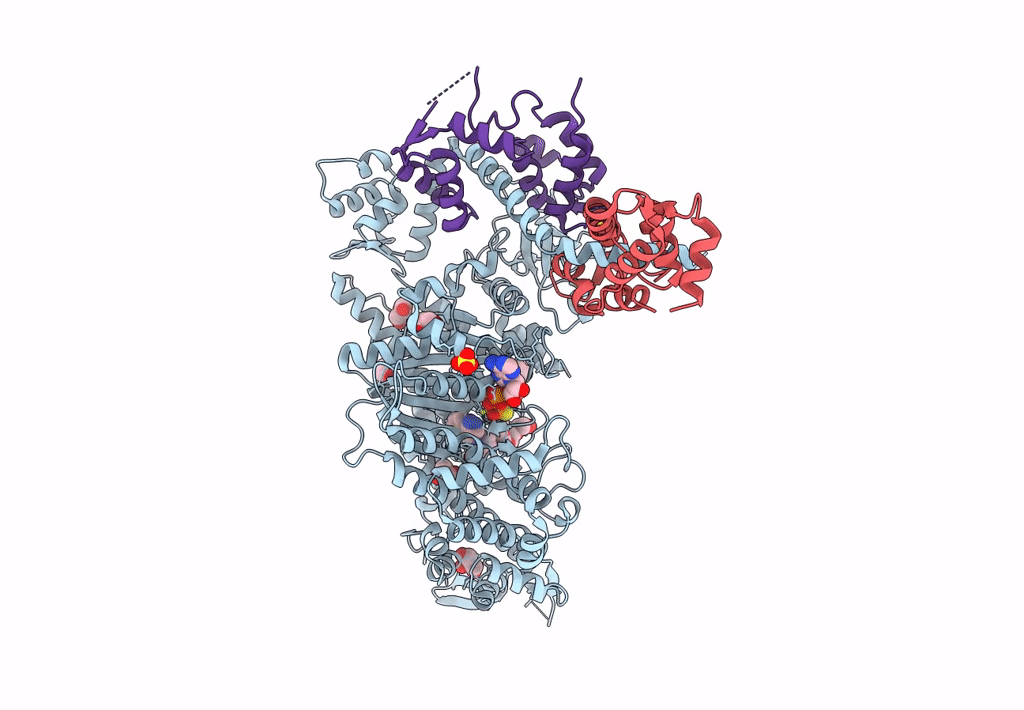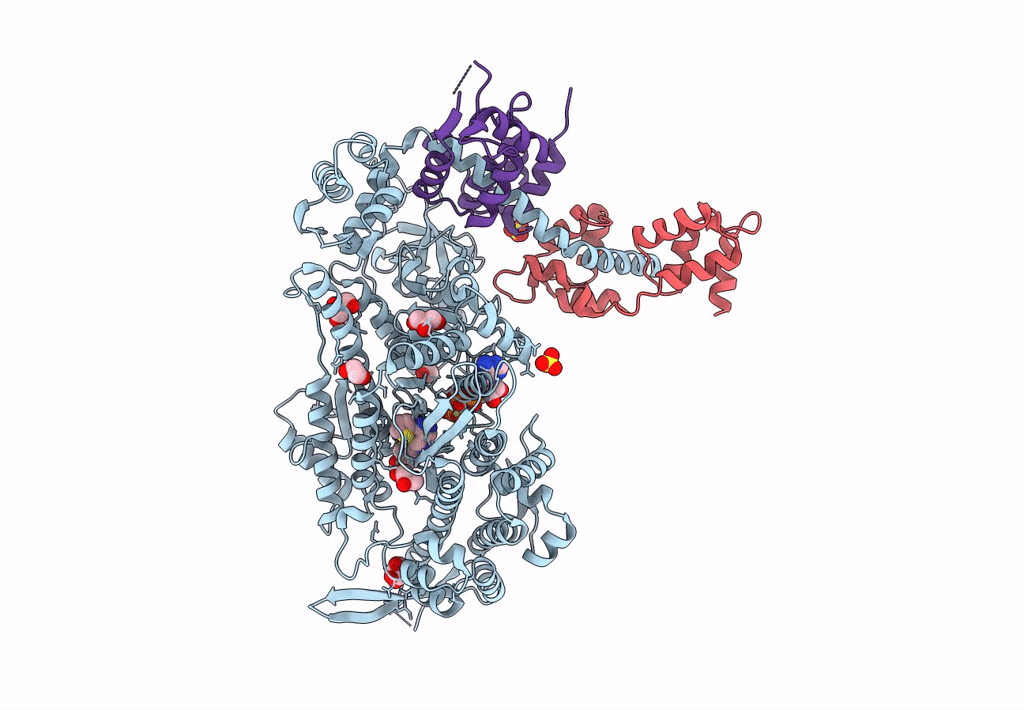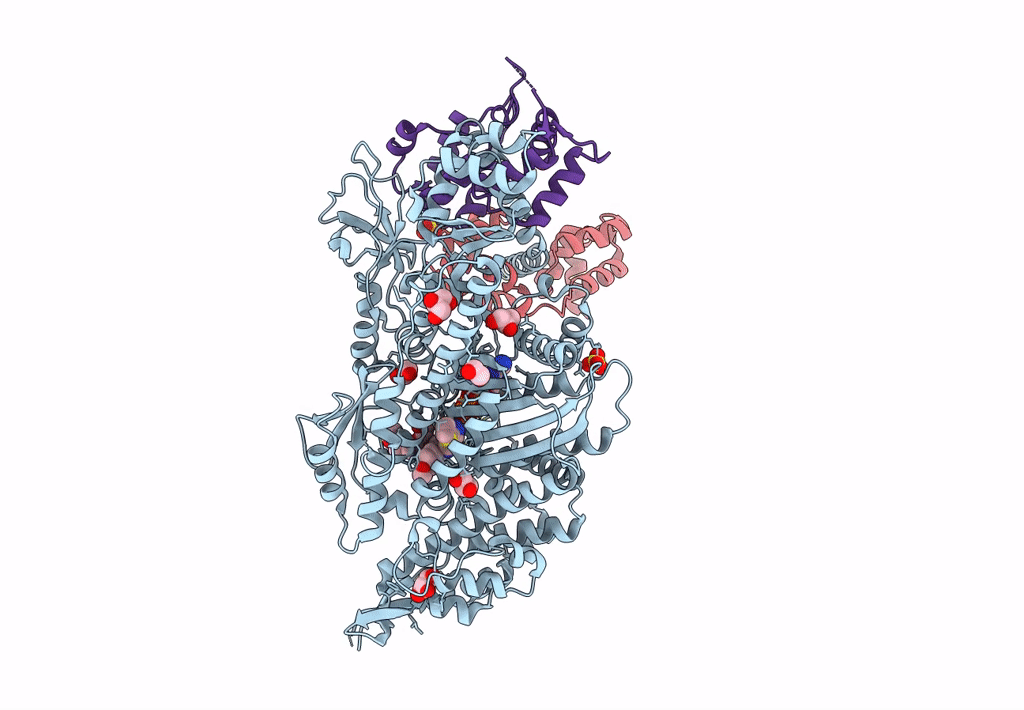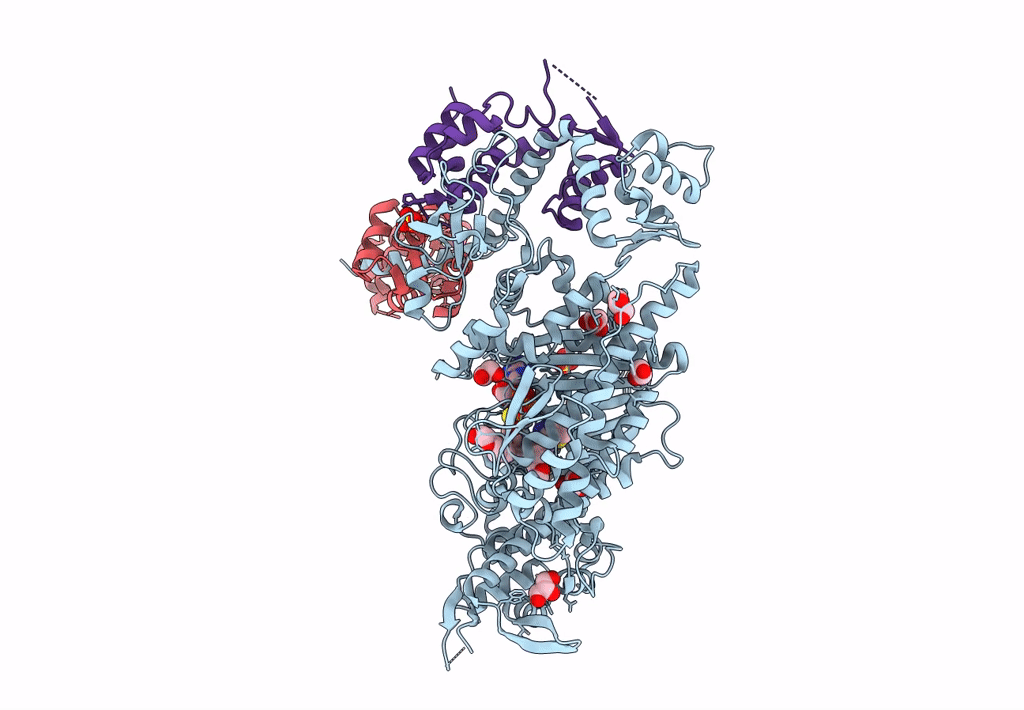
Plasmodium Falciparum Myosin A Full-Length, Post-Rigor State Complexed To Mg.Atp-Gamma-S
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-06-21 Classification: MOTOR PROTEIN Ligands: ADP, SO4, EDO, MG |
 |
Plasmodium Falciparum Myosin A Full-Length, Post-Rigor State Complexed To The Inhibitor Knx-002 And Mg.Atp-Gamma-S
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-06-21 Classification: MOTOR PROTEIN Ligands: GOL, SO4, EDO, PG4, AGS, MG, KQ0 |
 |
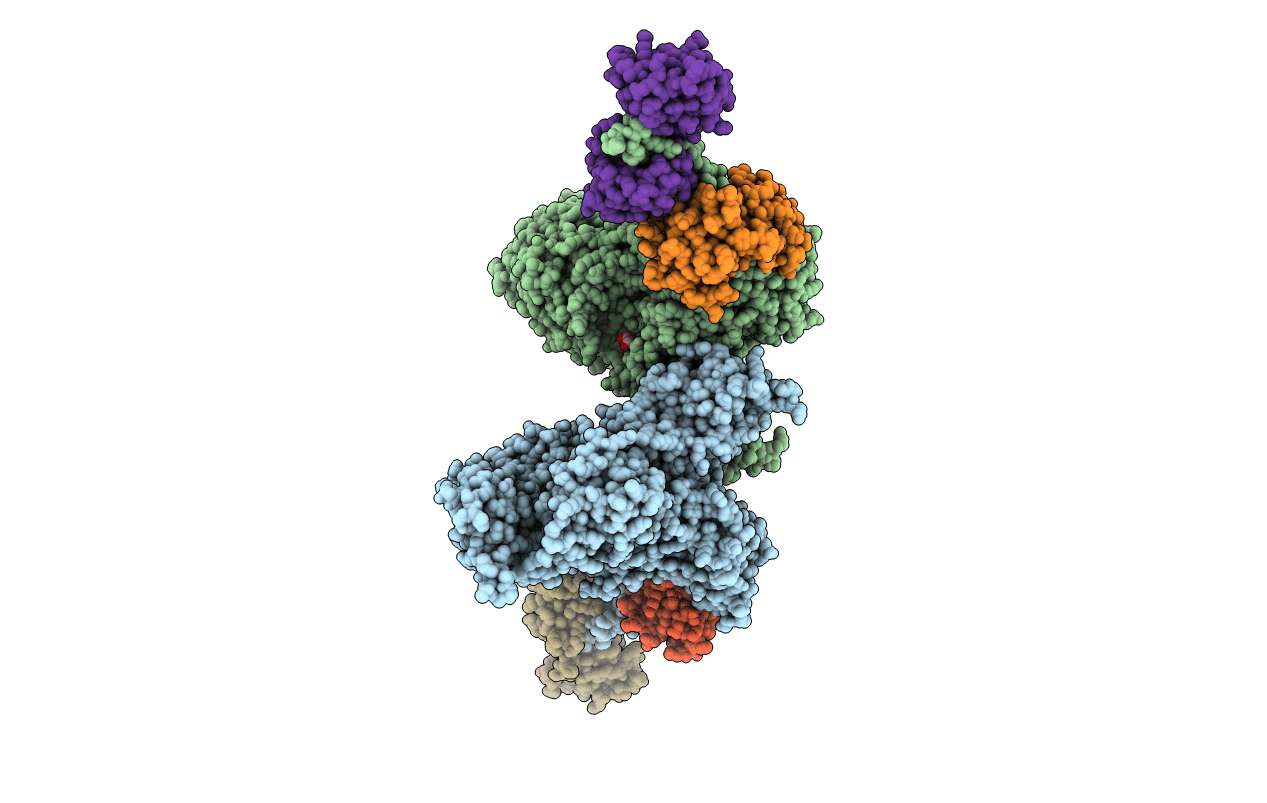
Cryo-Em Structure Of The Divergent Actomyosin Complex From Plasmodium Falciparum Myosin A In The Rigor State
Organism: Plasmodium falciparum (isolate 3d7)
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: MOTOR PROTEIN Ligands: ADP, MG, 9UE |
 |
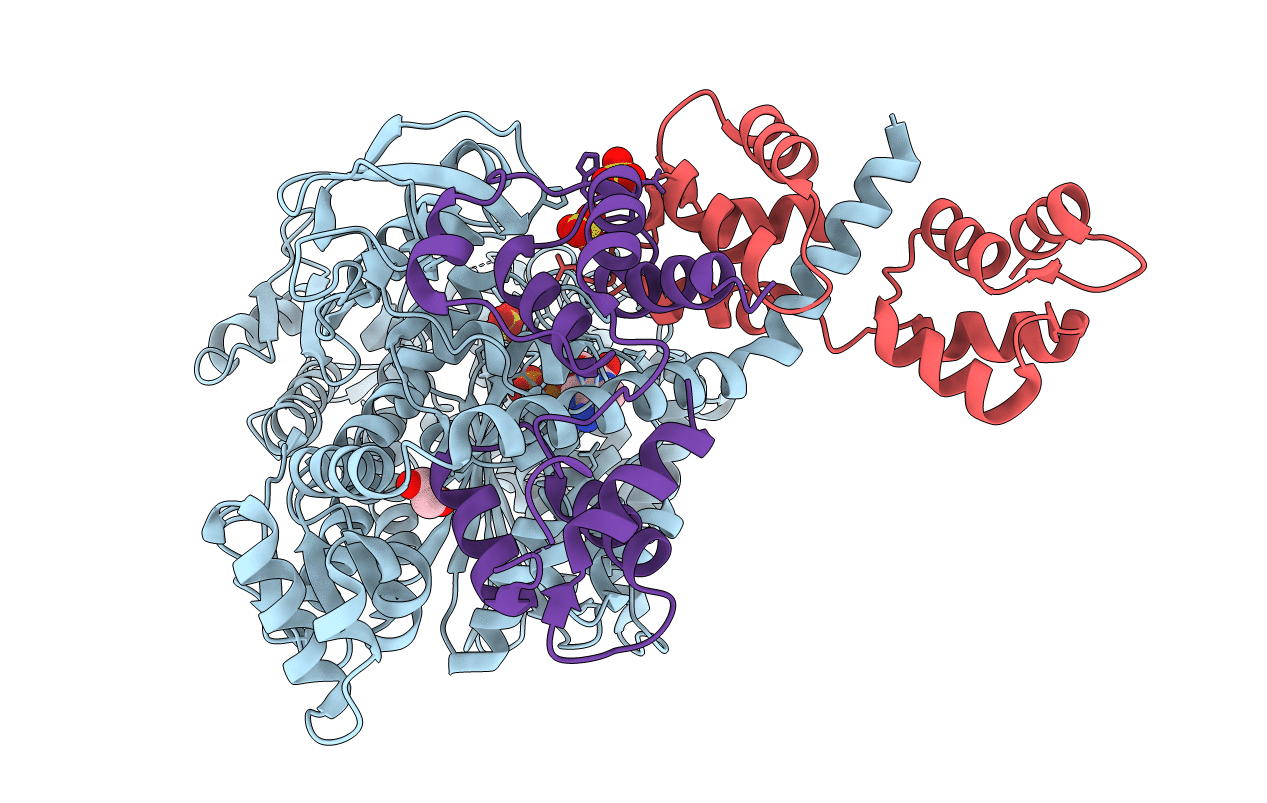
Organism: Plasmodium falciparum (isolate 3d7), Plasmodium falciparum (isolate nf54)
Method: X-RAY DIFFRACTION Resolution:3.99 Å Release Date: 2020-11-11 Classification: MOTOR PROTEIN Ligands: ADP, VO4, MG |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-11-11 Classification: MOTOR PROTEIN Ligands: EDO, SO4, MG, ADP |
 |
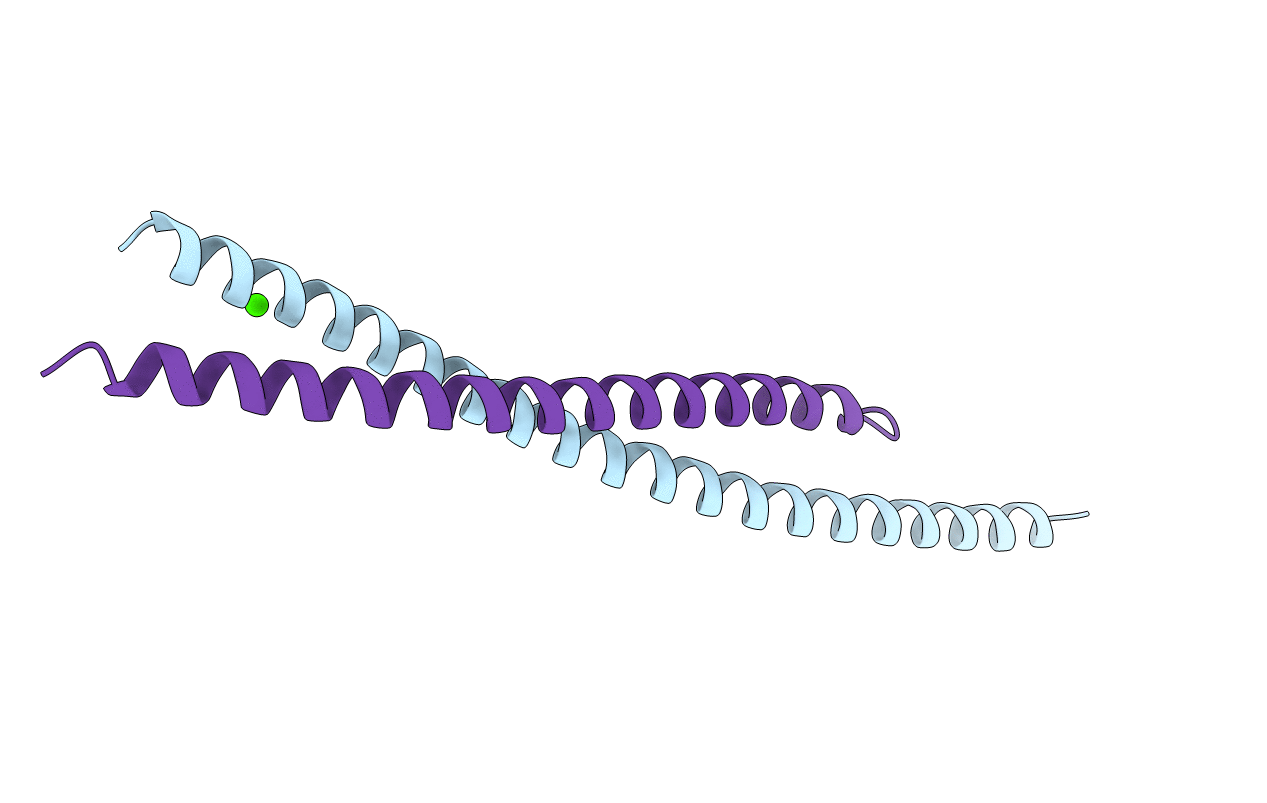
Organism: Plasmodium falciparum (isolate 3d7), Plasmodium falciparum (isolate nf54)
Method: X-RAY DIFFRACTION Resolution:3.27 Å Release Date: 2020-11-11 Classification: MOTOR PROTEIN Ligands: SO4, EDO, MG, ADP |
 |
Coiled-Coil Registry Shifts In The F684I Mutant Of Bicaudal D Result In Cargo-Independent Activation Of Dynein Motility
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-05-13 Classification: MOTOR PROTEIN Ligands: CA |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2019-08-07 Classification: MOTOR PROTEIN Ligands: EDO, GOL |
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2019-08-07 Classification: MOTOR PROTEIN Ligands: ADP, MG, VO4 |
 |
The Structure Of The Actin-Smooth Muscle Myosin Motor Domain Complex In The Rigor State
Organism: Gallus gallus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2018-09-19 Classification: MOTOR PROTEIN Ligands: ADP, MG |
 |
Em Structure Of The Heavy Meromyosin Subfragment Of Chick Smooth Muscle Myosin With Regulatory Light Chain In Phosphorylated State
Organism: Gallus gallus
Method: ELECTRON MICROSCOPY Release Date: 2011-11-16 Classification: STRUCTURAL PROTEIN |
 |
Structures Of Actin-Bound Wh2 Domains Of Spire And The Implication For Filament Nucleation
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-06-02 Classification: Contractile Protein/Protein binding Ligands: CA, ATP |
 |
Structures Of Actin-Bound Wh2 Domains Of Spire And The Implication For Filament Nucleation
Organism: Drosophila melanogaster, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-06-02 Classification: Contractile Protein/Protein binding Ligands: CA, ATP, LAB |
 |
Structures Of Actin-Bound Wh2 Domains Of Spire And The Implication For Filament Nucleation
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-06-02 Classification: Contractile Protein/Protein binding Ligands: CA, ATP |
 |
Structures Of Actin-Bound Wh2 Domains Of Spire And The Implication For Filament Nucleation
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-05-26 Classification: Contractile Protein/Protein binding Ligands: ATP, CA |
 |
Structures Of Actin-Bound Wh2 Domains Of Spire And The Implication For Filament Nucleation
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-05-26 Classification: Contractile Protein/Protein binding Ligands: ATP, CA |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-10-07 Classification: CONTRACTILE PROTEIN Ligands: ATP, CA, CY9 |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-10-07 Classification: CONTRACTILE PROTEIN Ligands: ATP, CA, CY9 |

