Search Count: 45
 |
Organism: Mycobacterium tuberculosis cdc1551
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: NAD, 53K, CL, NA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: NAD, XT3 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: NAD, XT0, CL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: NAD, XT5, CL, NA |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: NAD, XTW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: NAD, XTV |
 |
Crystal Structure Of Escherichia Coli Menb In Complex With Substrate Analogue, Osb-Ncoa
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-24 Classification: LYASE/LYASE INHIBITOR Ligands: S0N, CL, PGE, GOL |
 |
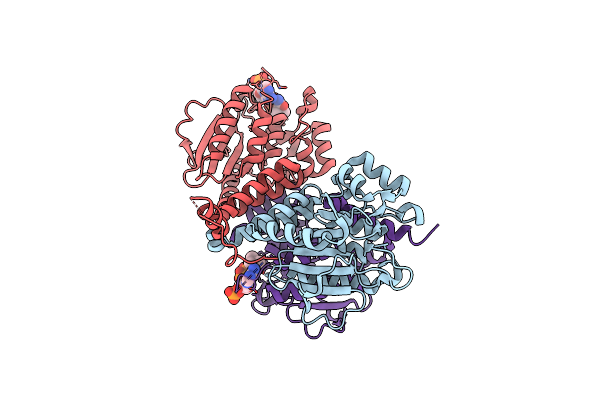
Crystal Structure Of Escherichia Coli Menb, The 1,4-Dihydroxy-2-Naphthoyl-Coa Synthase In Vitamin K2 Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-08-24 Classification: LYASE Ligands: MLI, GOL |
 |
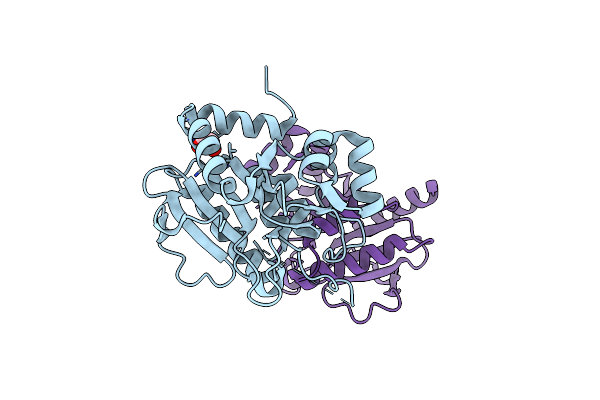
Crystal Structure Of Mycobacterium Tuberculosis Menb In Complex With Substrate Analogue, Osb-Ncoa
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-08-24 Classification: LYASE/LYASE INHIBITOR Ligands: S0N |
 |
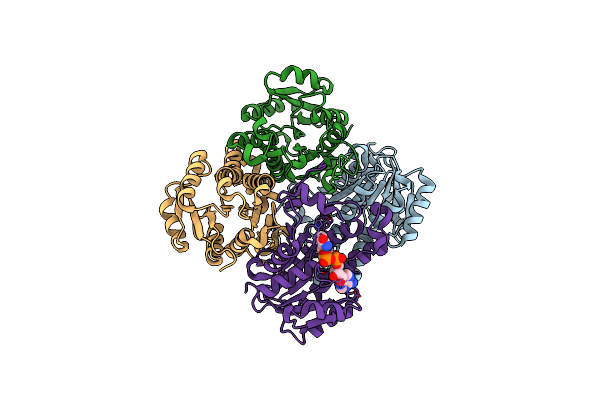
Crystal Structure Of Mycobacterium Tuberculosis Menb With Altered Hexameric Assembly
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-08-24 Classification: LYASE Ligands: GOL |
 |
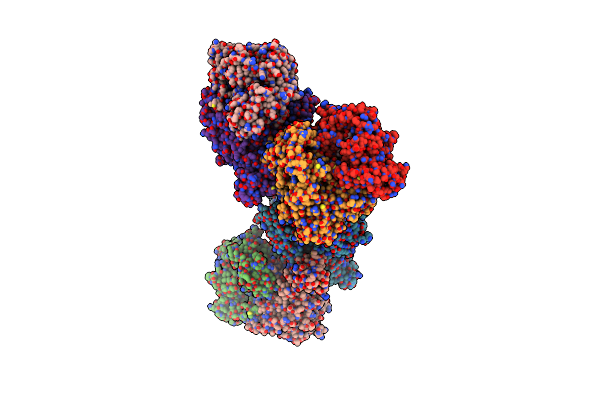
Crystal Structure Of Francisella Tularensis Enoyl Reductase (Ftfabi) With Bound Nad
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2009-02-24 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Xanthine Dehydrogenase (Desulfo Form) From Rhodobacter Capsulatus In Complex With Hypoxanthine
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, MTE, CA, HPA, MOM |
 |
Crystal Structure Of Xanthine Dehydrogenase (Desulfo Form) From Rhodobacter Capsulatus In Complex With Xanthine
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, MTE, CA, XAN, MOM |
 |
Crystal Structure Of Xanthine Dehydrogenase From Rhodobacter Capsulatus In Complex With Bound Inhibitor Pterin-6-Aldehyde
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, XAX, BA, HHR |
 |
Crystal Structure Of Xanthine Dehydrogenase (E232Q Variant) From Rhodobacter Capsulatus In Complex With Hypoxanthine
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2008-12-23 Classification: OXIDOREDUCTASE Ligands: FES, FAD, XAX, BA, HPA |
 |
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-07-08 Classification: HYDROLASE Ligands: SF4, CA |
 |
Crystal Structure Of Cell Division Protein Ftsz From Mycobacterium Tuberculosis In Complex With Citrate.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-05-06 Classification: Cell Cycle, Signaling Protein Ligands: CIT |
 |
Crystal Structure Of Cell Division Protein Ftsz From Mycobacterium Tuberculosis In Complex With Gtp-Gamma-S
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-05-06 Classification: Cell Cycle, Signaling Protein Ligands: GSP |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2007-02-06 Classification: HYDROLASE |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-02-06 Classification: HYDROLASE Ligands: CL |

