Search Count: 121
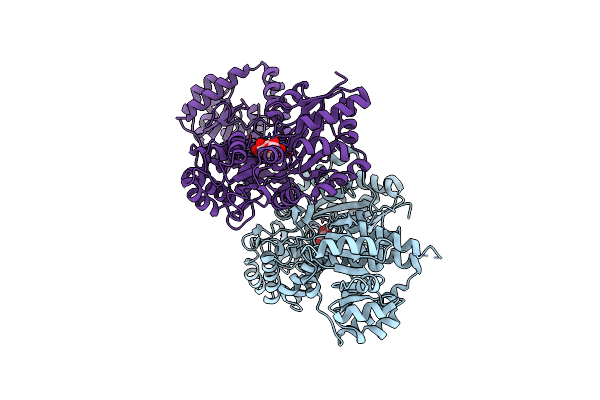 |
Crystal Structure Of Mbp Fusion With Hppk From Methanocaldococcus Jannaschii
Organism: Escherichia coli k-12, Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: TRANSFERASE |
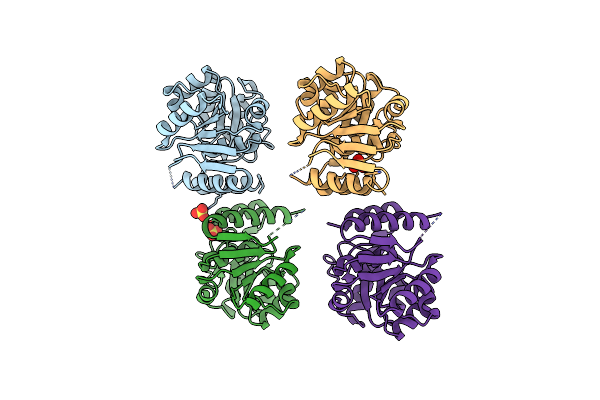 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: SO4 |
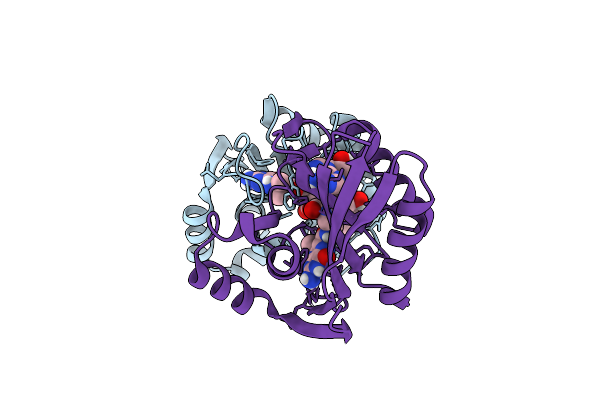 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-03 Classification: TRANSFERASE/INHIBITOR Ligands: H73 |
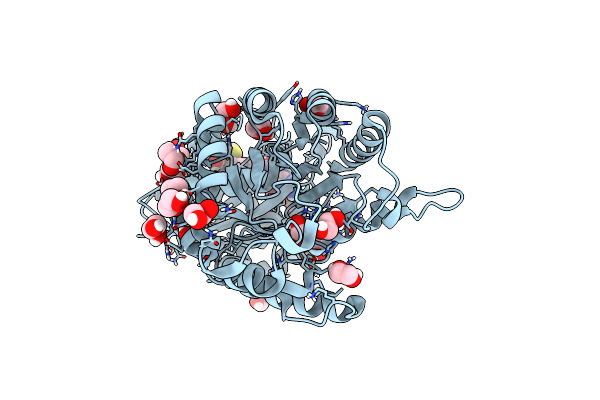 |
Crystal Structure Of Francisella Tularensis Hppk-Dhps In Complex With Hppk Inhibitor Hp-73
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: H73, EDO, CL |
 |
Crystal Structure Of Clostridioides Difficile Protein Tyrosine Phosphatase At Ph 8.5
Organism: Clostridioides difficile 6050
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-07-05 Classification: HYDROLASE |
 |
Crystal Structure Of Clostridioides Difficile Protein Tyrosine Phosphatase At Ph 7.5
Organism: Clostridioides difficile 6050
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-07-05 Classification: HYDROLASE Ligands: EPE, CL |
 |
Crystal Structure Of Yersinia Pestis Dihydrofolate Reductase At 1.25-A Resolution
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: SO4, CL, MG |
 |
Crystal Structure Of Yersinia Pestis Dihydrofolate Reductase In Complex With Trimethoprim
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: TOP, SO4, EDO, CL |
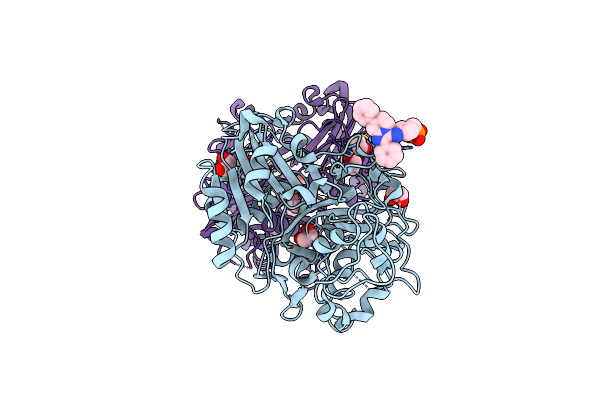 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: N8I, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: N7U, EDO, DMS |
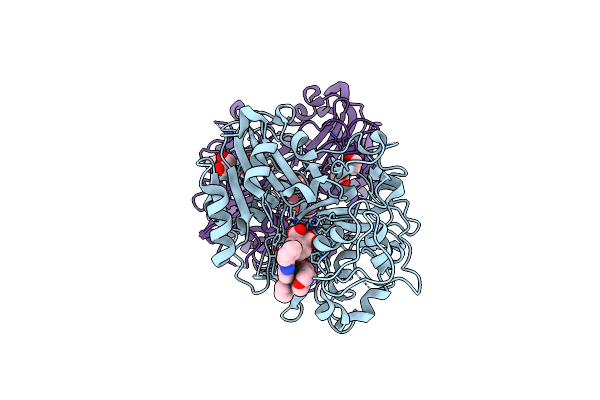 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: OYH, MPO, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: OYR, DMS, MPO, EDO |
 |
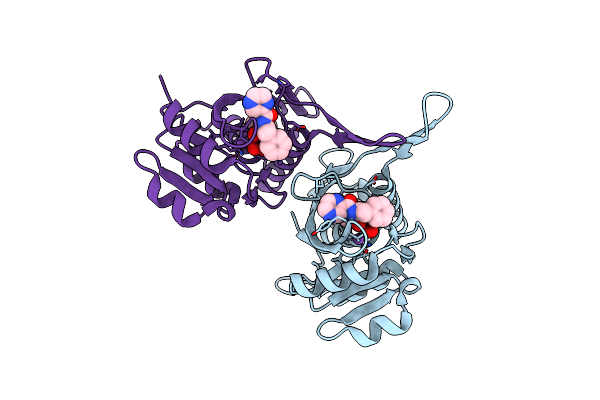
Organism: Helicobacter pylori g27
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-03-01 Classification: LYASE Ligands: PE0, EDO |
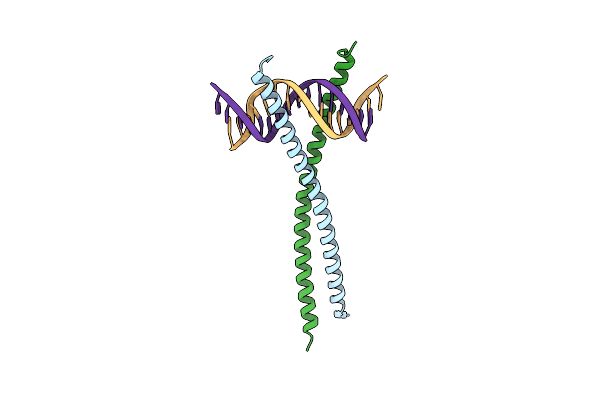 |
Structural Basis For Cell Type Specific Dna Binding Of C/Ebpbeta: The Case Of Cell Cycle Inhibitor P15Ink4B Promoter
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Atp-Dependent Lon Protease From Bacillus Subtillis (Bslonba)
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: BO2, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-03-24 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TG7, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-03-24 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TGV, EDO |
 |
Organism: Streptomyces sviceus atcc 29083
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2021-03-17 Classification: FLAVOPROTEIN Ligands: FAD, NAP |
 |
Organism: Streptomyces sviceus atcc 29083
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2021-03-17 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2019-11-20 Classification: HYDROLASE Ligands: CL |

