Search Count: 17
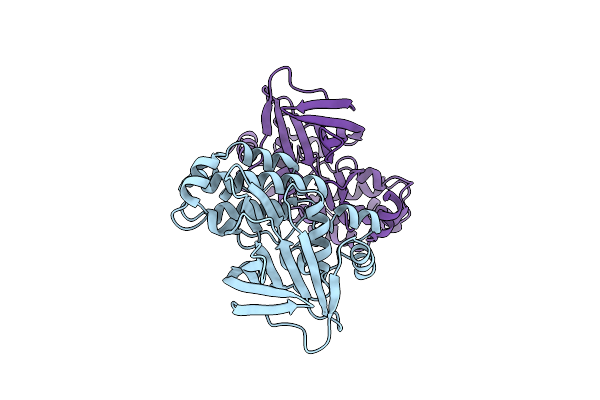 |
Crystal Structure Of The Kinetoplastid Kinetochore Protein Kkt14 C-Terminal Domain From Apiculatamorpha Spiralis
Organism: Apiculatamorpha spiralis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-17 Classification: CELL CYCLE |
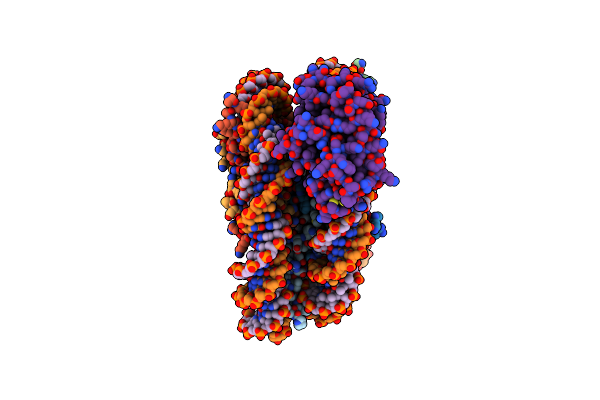 |
Organism: Saccharomyces cerevisiae, Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
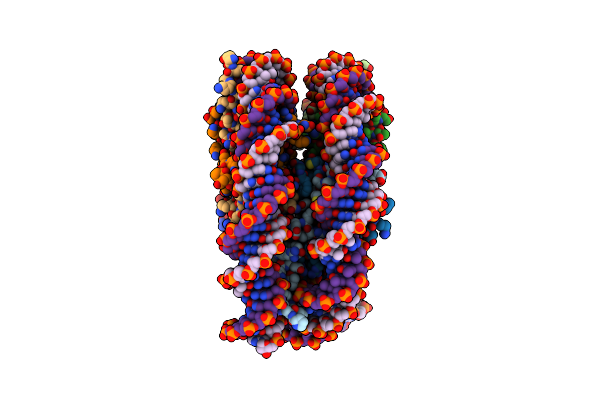 |
Organism: Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: DNA BINDING PROTEIN/DNA |
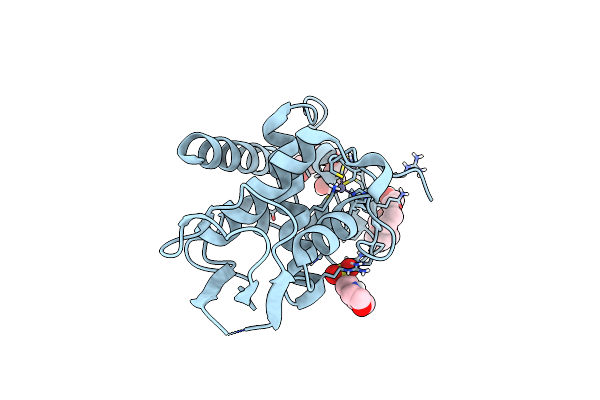 |
Organism: Vanderwaltozyma polyspora dsm 70294
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-29 Classification: DNA BINDING PROTEIN Ligands: ZN, MES, 1PE, PG4 |
 |
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-11-07 Classification: CELL CYCLE |
 |
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-11-07 Classification: CELL CYCLE |
 |
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-11-07 Classification: CELL CYCLE |
 |
Organism: Candida glabrata, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-07 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-24 Classification: TRANSFERASE Ligands: PG4, GOL, PGE, MLI, PEG |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-08-25 Classification: LYASE |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-08-25 Classification: LYASE Ligands: 5RP |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-25 Classification: LYASE Ligands: MG |
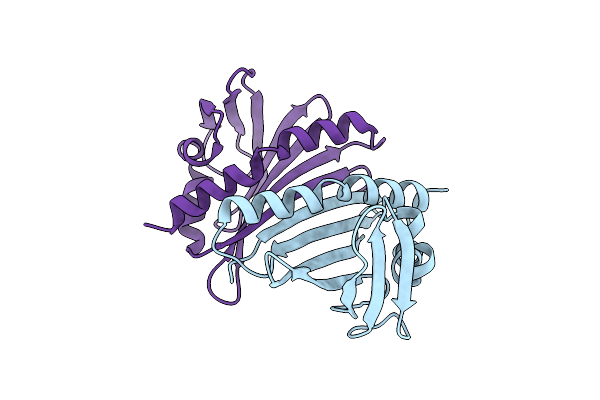 |
Structural Characterization Of Smu.440, A Hypothetical Protein From Streptococcus Mutans
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2009-08-25 Classification: UNKNOWN FUNCTION |
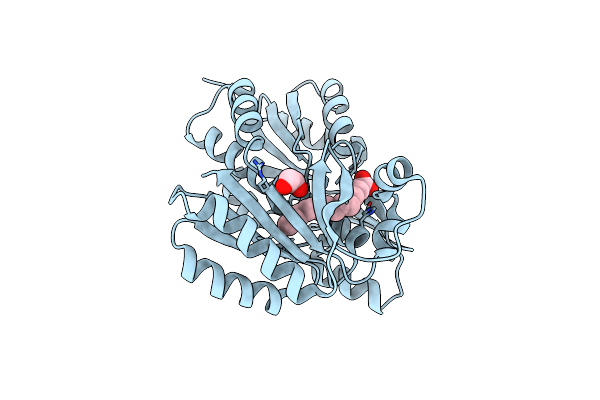 |
Crystal Structure Of Degv, A Fatty Acid Binding Protein From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-05-12 Classification: FATTY ACID-BINDING PROTEIN Ligands: PLM, EDO, BR |
 |
Crystal Structure Of A Thermostable Serine Protease Al20 From Extremophilic Microoganism
Organism: Nesterenkonia abyssinica
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2009-02-10 Classification: HYDROLASE Ligands: FMT |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-26 Classification: HYDROLASE Ligands: ACT, ZN |
 |
Crystal Structure Of Imidazolonepropionase Complexed With Imidazole-4-Acetic Acid Sodium Salt, A Substrate Homologue
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-26 Classification: HYDROLASE Ligands: ZN, IZC |

