Search Count: 19
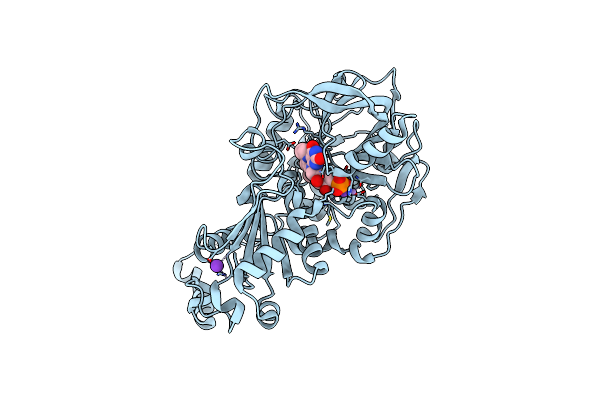 |
Structure Of A. Niger Fdc1 With The Prenylated-Flavin Cofactor In The Iminium Form.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, MN, K |
 |
Structure Of A. Niger Fdc1 With The Prenylated-Flavin Cofactor In The Iminium And Ketimine Forms.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K |
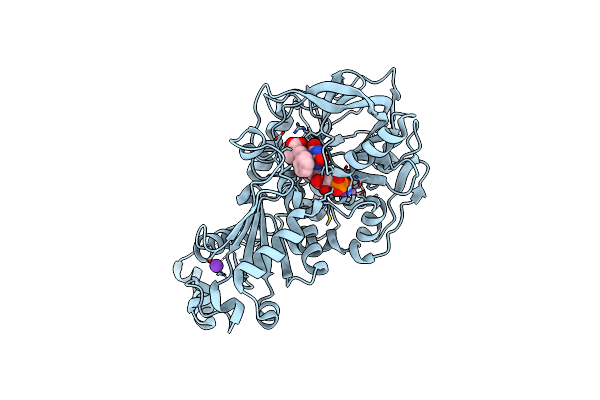 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, 4LV |
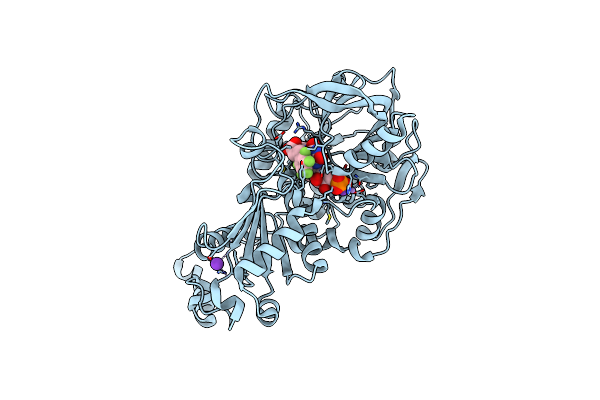 |
Crystal Structure Of A Niger Fdc1 In Complex With Penta-Fluorocinnamic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, F5C |
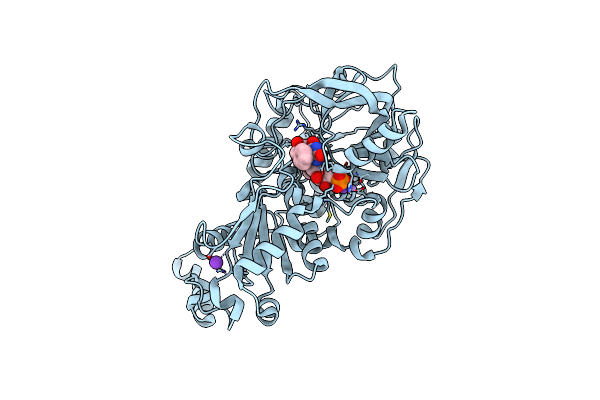 |
Structure Of A. Niger Fdc1 In Complex With A Phenylpyruvate Derived Adduct To The Prenylated Flavin Cofactor
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4MJ, MN, K |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, 4M4, CO2 |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, FZZ, MN, K, 4LW |
 |
Structure Of S. Cerevisiae Fdc1 With The Prenylated-Flavin Cofactor In The Iminium Form.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-06-17 Classification: LYASE |
 |
Structure Of C. Dubliensis Fdc1 With The Prenylated-Flavin Cofactor In The Iminium Form.
Organism: Candida dubliniensis
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2015-06-17 Classification: LYASE Ligands: MN, K, 4LU |
 |
Structure Of Ubix In Complex With Oxidised Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LR, FMN, SCN, K |
 |
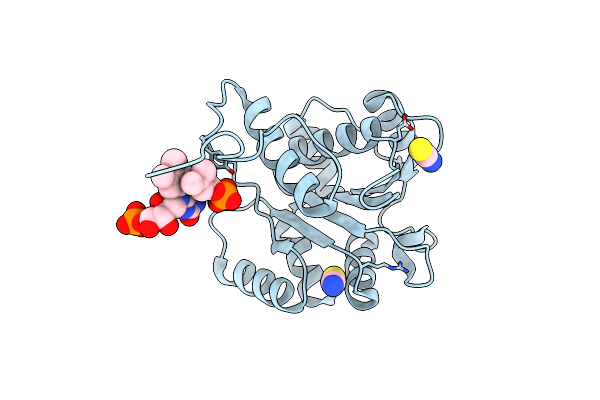
Structure Of Ubix E49Q Mutant In Complex With Oxidised Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LR, FMN, SCN, K |
 |
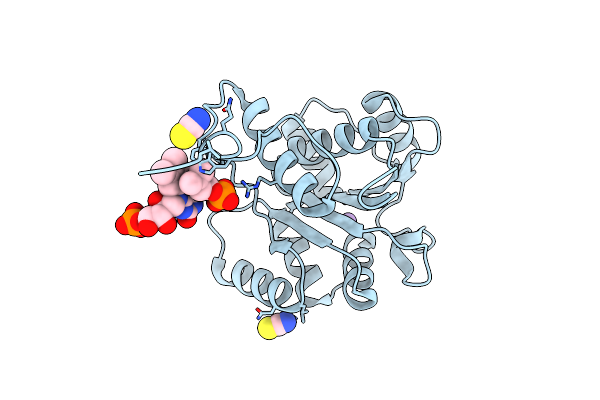
Structure Of Ubix E49Q Mutant In Complex With Reduced Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LR, FNR, SCN |
 |
Structure Of Ubix Y169F In Complex With Oxidised Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LR, FMN, SCN, K |
 |
Ubix In Complex With A Covalent Adduct Between Dimethylallyl Monophosphate And Reduced Fmn
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LS, SCN, NA, PO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, SCN, PO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2015-06-17 Classification: TRANSFERASE Ligands: 4LU, SCN, PO4 |
 |
Structure Of Ubix E49Q In Complex With A Covalent Adduct Between Dimethylallyl Monophosphate And Reduced Fmn
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LS, SCN, K, PO4 |
 |
Structure Of Ubix Y169F In Complex With A Covalent Adduct Formed Between Reduced Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LS, SCN, NA, PO4 |
 |
Organism: Piriformospora indica
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-05-15 Classification: ISOMERASE Ligands: K, NA, PO4 |

