Search Count: 41
 |
Cryoem Structure Of Mammalian Aap In Complex With Acetyl-Alanyl-Chloromethylketone
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: A1INA |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: A1ING |
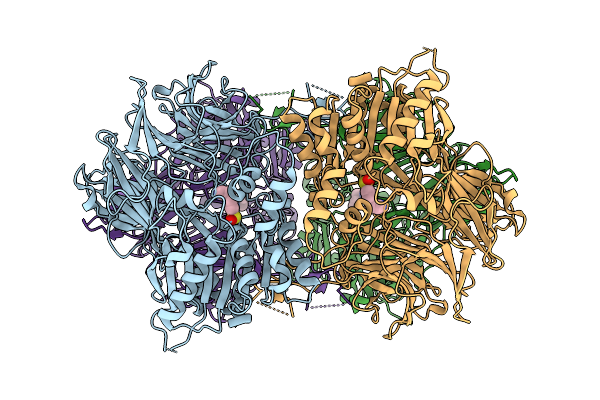 |
Cryo-Em Structure Of Acylaminoacyl Peptidase (Aap) In Covalent Complex With Inhibitor Aebsf
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: AES |
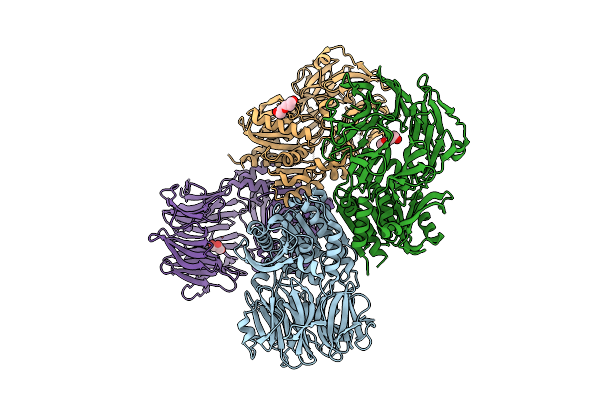 |
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: HYDROLASE Ligands: 1AX |
 |
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRUS |
 |
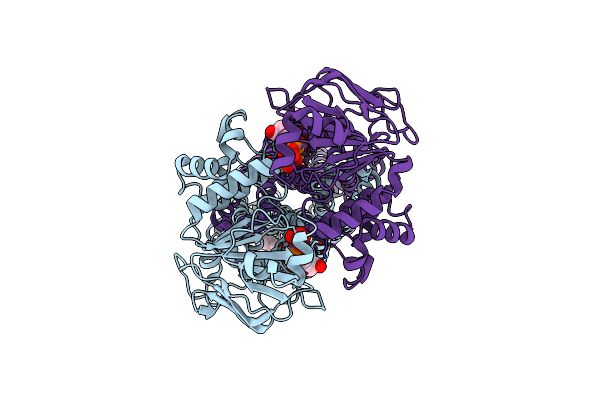
Organism: Rhodobacter capsulatus sb 1003
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRUS |
 |
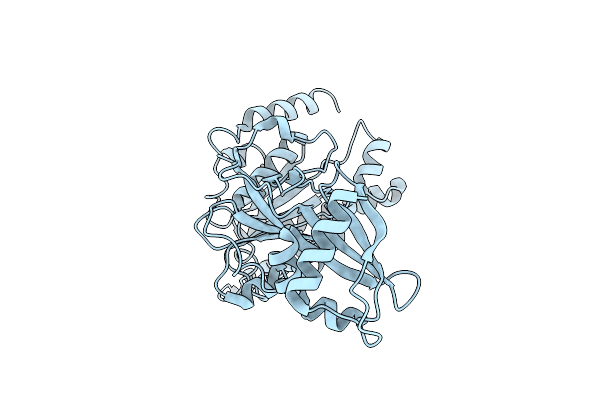
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: MEMBRANE PROTEIN Ligands: VO4, ADP, MG, POV |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-03-04 Classification: TRANSLOCASE |
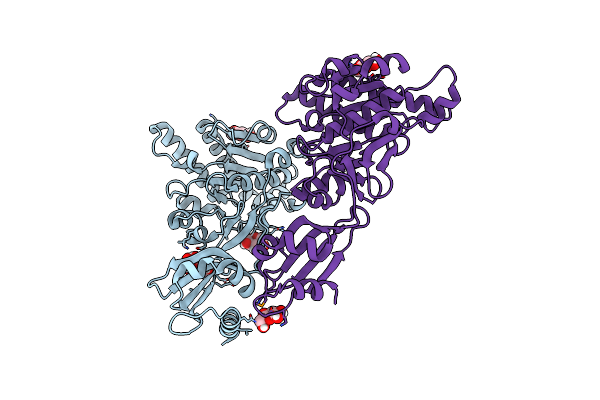 |
Structure Of Pcw3 Conjugation Coupling Protein Tcpa Monomer Orthorhombic Crystal Form
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-26 Classification: TRANSLOCASE Ligands: BGC |
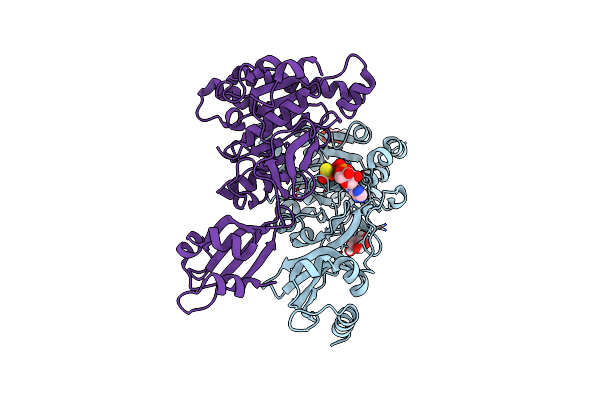 |
Structure Of Pcw3 Conjugation Coupling Protein Tcpa Monomer Form With Atpgs
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2020-02-26 Classification: TRANSLOCASE Ligands: BGC, AGS |
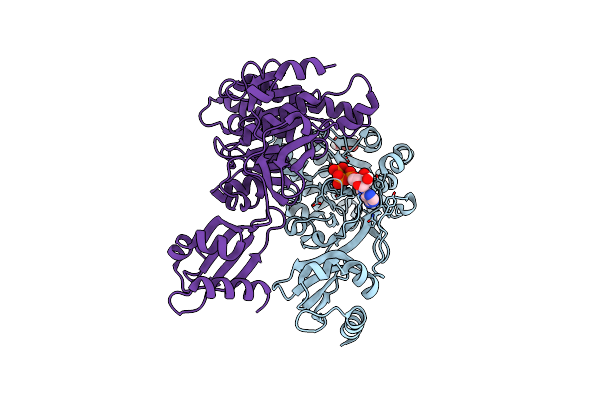 |
Structure Of Pcw3 Conjugation Coupling Protein Tcpa Monomeric Form With Atp
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-02-26 Classification: TRANSLOCASE Ligands: ATP, BGC |
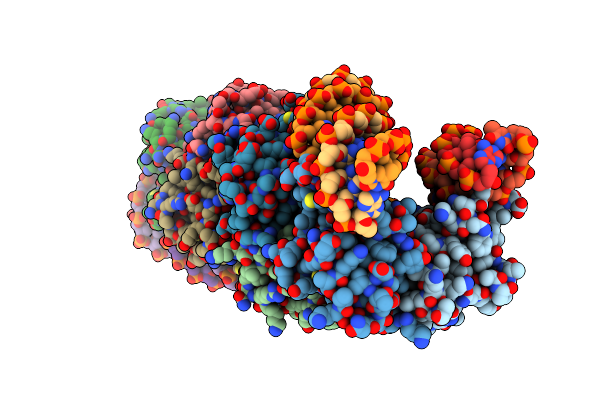 |
Structure Of An Accessory Protein Of The Pcw3 Relaxosome In Complex With The Origin Of Transfer (Orit) Dna
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2018-04-18 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2018-04-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-23 Classification: SIGNALING PROTEIN Ligands: MLA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-09-23 Classification: Signaling Protein/Inhibitor |
 |
Crystal Structure Of Phanta, A Weakly Fluorescent Photochromic Gfp-Like Protein. On State
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-08 Classification: FLUORESCENT PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-08 Classification: FLUORESCENT PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-04-08 Classification: FLUORESCENT PROTEIN |
 |
Structure Of Membrane Binding Protein Pleurotolysin A From Pleurotus Ostreatus
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-02-18 Classification: MEMBRANE BINDING PROTEIN Ligands: SO4 |
 |
Structure Of Membrane Binding Protein Pleurotolysin B From Pleurotus Ostreatus
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-02-18 Classification: MEMBRANE BINDING PROTEIN Ligands: ACT, GOL, CL |

