Search Count: 64,177
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: A1AWG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: PROTEIN BINDING Ligands: GOL, A1IU1, DMS |
 |
Crystal Structure Of Phosphatidyl Inositol 4-Kinase Ii Beta In Complex With Hh5129
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: A1IVA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: PROTEIN BINDING Ligands: GOL, A1IV7, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: PROTEIN BINDING Ligands: HHQ, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: PROTEIN BINDING Ligands: GOL, LL0, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: PROTEIN BINDING Ligands: GOL, UJK, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: PROTEIN BINDING Ligands: UUG, DMS |
 |
Crystal Structure Of Zika Virus Ns2B-Ns3 Protease In Complex With Compound 2
Organism: Zika virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: A1I16, ACY |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1D7W |
 |
Structure Of Human Neuronal Nitric Oxide Synthase R354A/G357D Mutant Heme Domain Bound With 7-(3-Aminomethyl)Phenyl-6-Fluoro-4-Methylquinoin-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BUD, GOL, ZN |
 |
Structure Of Human Neuronal Nitric Oxide Synthase R354A/G357D Mutant Heme Domain Bound With 2-(2-Amino-6-Fluoro-4-Methylquinolin-7-Yl)-5-(Aminomethyl)Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BUF, GOL, ZN |
 |
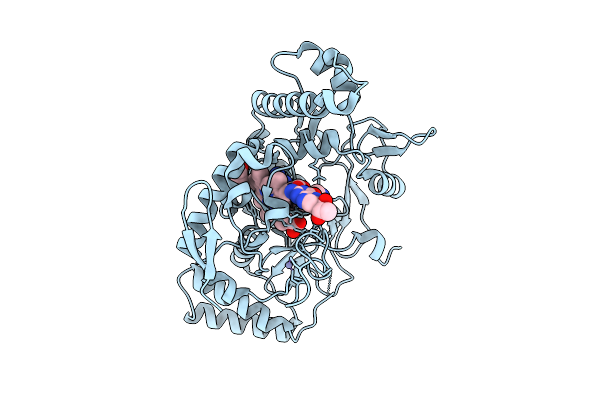
Structure Of Human Neuronal Nitric Oxide Synthase R354A/G357D Mutant Heme Domain Bound 2-(2-Amino-6-Fluoro-4-Methylquinolin-7-Yl)-5-(2-Aminoethyl)Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BUH, GOL, ZN |
 |
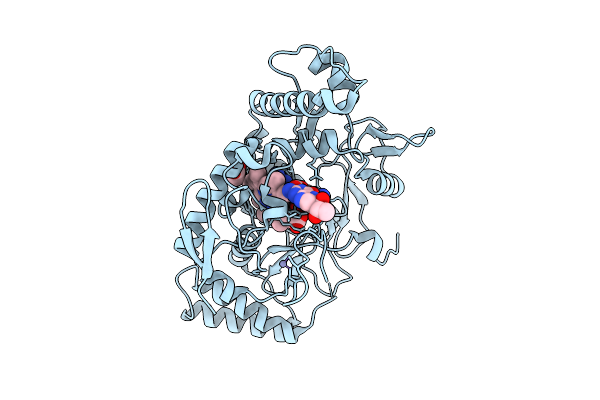
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 7-(3-Aminomethyl)Phenyl-6-Fluoro-4-Methylquinoin-2-Amine
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BUD, ACT, ZN |
 |
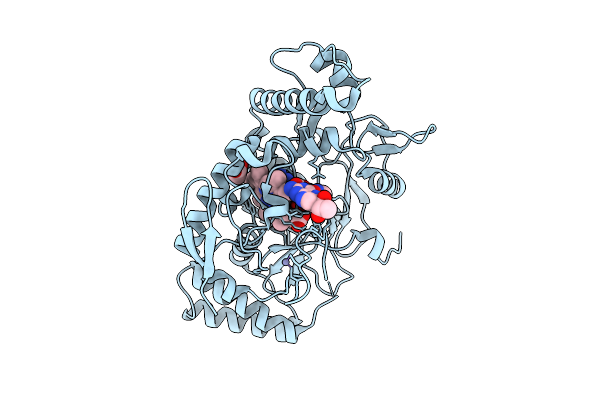
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound With 2-(2-Amino-6-Fluoro-4-Methylquinolin-7-Yl)-5-(Aminomethyl)Phenol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ACT, ZN, A1BUF |
 |
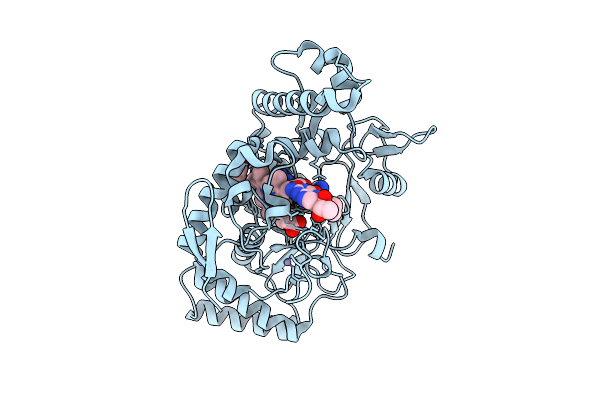
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound 2-(2-Amino-6-Fluoro-4-Methylquinolin-7-Yl)-5-(2-Aminoethyl)Phenol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, ACT, ZN, A1BUH |
 |
Structure Of Rat Neuronal Nitric Oxide Synthase R349A Mutant Heme Domain Bound 7-(3-(Aminomethyl)-4-(Cyclopropylmethoxy)Phenyl)-6-Fluoro-4-Methylquinolin-2-Amine
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BU3, ACT, ZN |
 |
Structure Of Human Endothelial Nitric Oxide Synthase Heme Domain Bound With 7-(3-Aminomethyl)Phenyl-6-Fluoro-4-Methylquinoin-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BUD, BTB, GOL, CL, GD, ZN |
 |
Structure Of Human Endothelial Nitric Oxide Synthase Heme Domain Bound With 2-(2-Amino-6-Fluoro-4-Methylquinolin-7-Yl)-5-(Aminomethyl)Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: HEM, H4B, A1BUF, BTB, GOL, CL, GD, ZN, CA |

