Search Count: 110
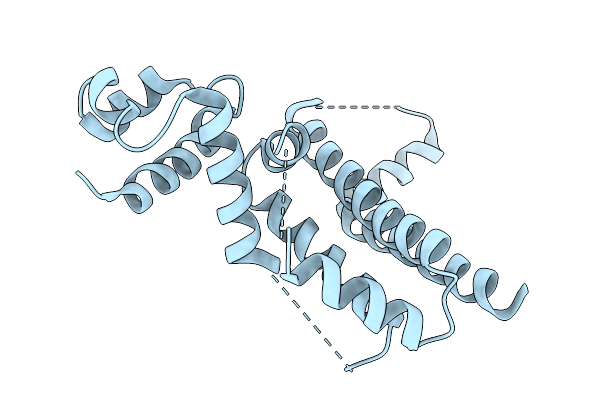 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L7-D1), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
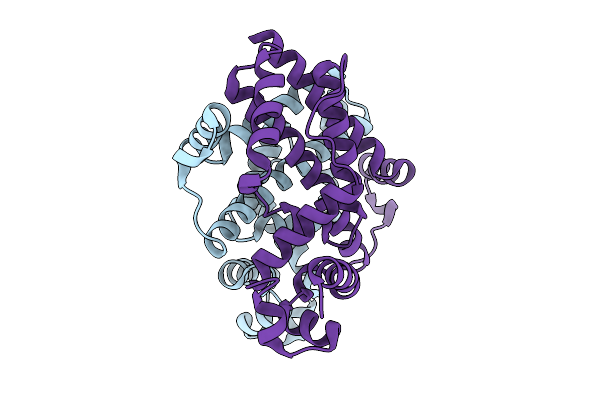 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
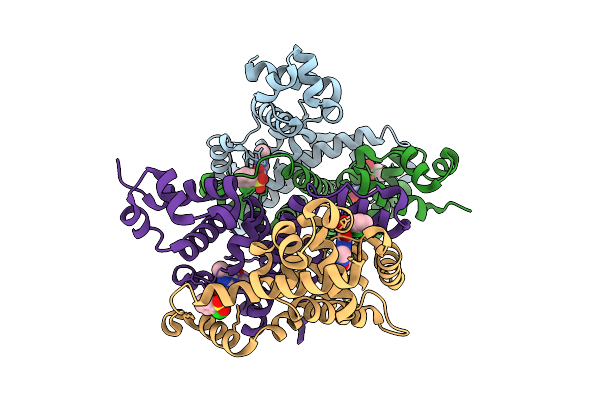 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Bound To Chlorsulfuron
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: 1CS |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN/DNA |
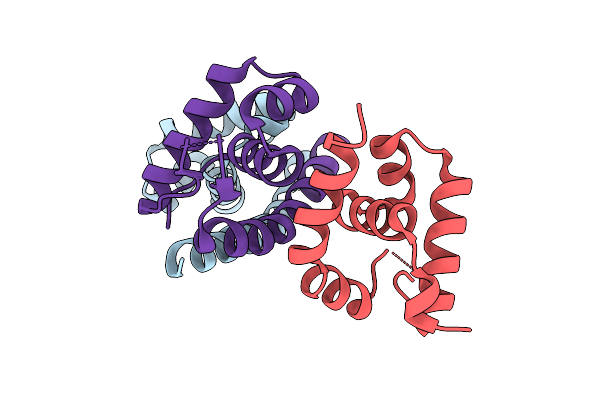 |
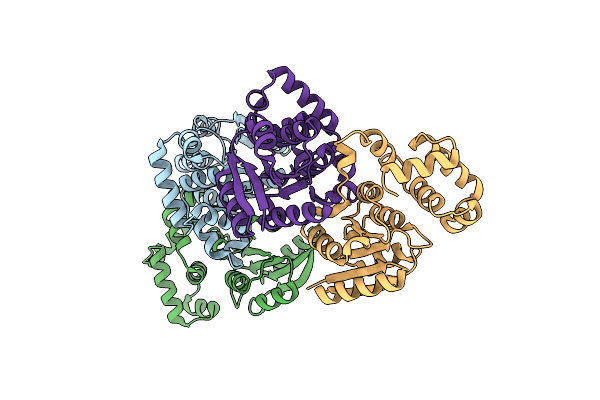
Crystal Structure Of The Acinetobacter Baumannii Lysr Family Regulator Acer Dna-Binding Domain (P321)
Organism: Acinetobacter baumannii (strain atcc 17978 / dsm 105126 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Dna-Binding Transcriptional Activator Evga From Escherichia Coli Str. K-12 Substr. Mg1655
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4, BOG |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2024-04-24 Classification: DNA BINDING PROTEIN/DNA |
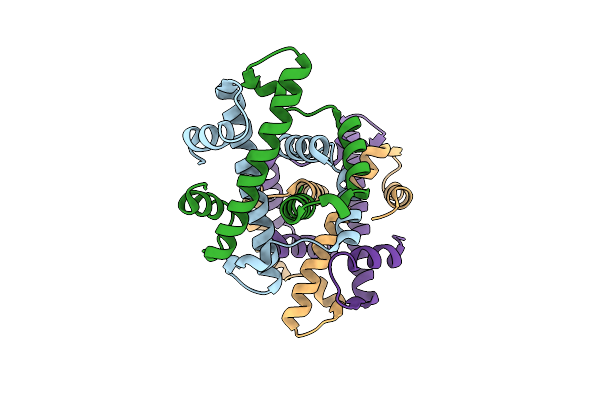 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN |
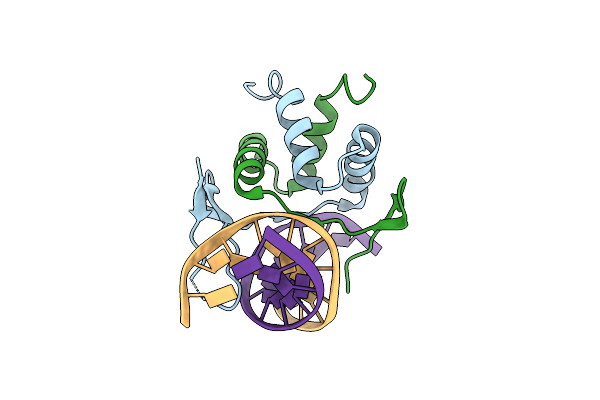 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-10-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-10-19 Classification: DNA BINDING PROTEIN |
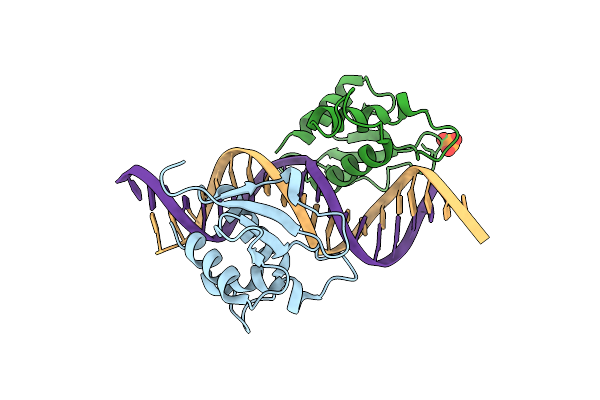 |
X-Ray Structure Of Interferon Regulatory Factor 4 Dna Binding Domain Bound To An Interferon-Stimulated Response Element
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-08 Classification: DNA BINDING PROTEIN Ligands: PO4 |
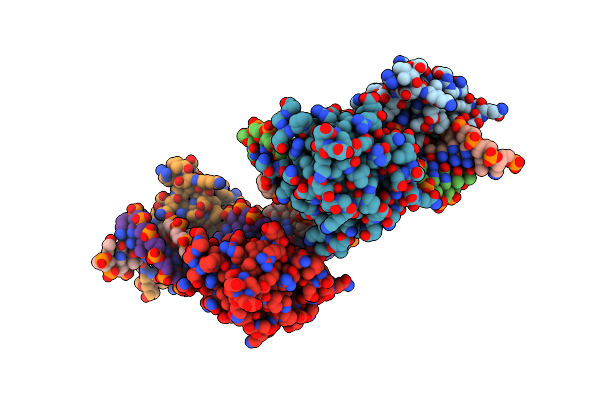 |
X-Ray Structure Of Interferon Regulatory Factor 4 Dna Binding Domain Bound To An Interferon-Stimulated Response Element
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2022-06-01 Classification: DNA BINDING PROTEIN |
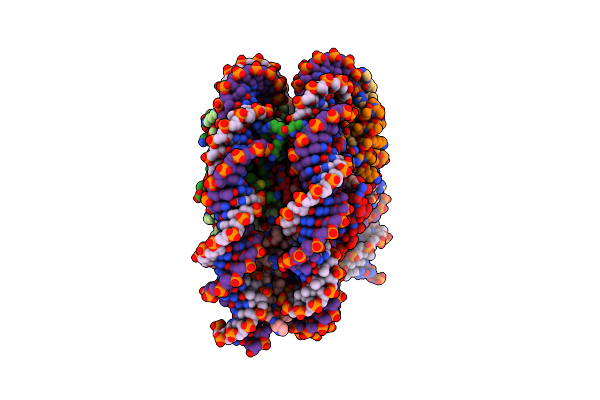 |
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: DNA BINDING PROTEIN/DNA |
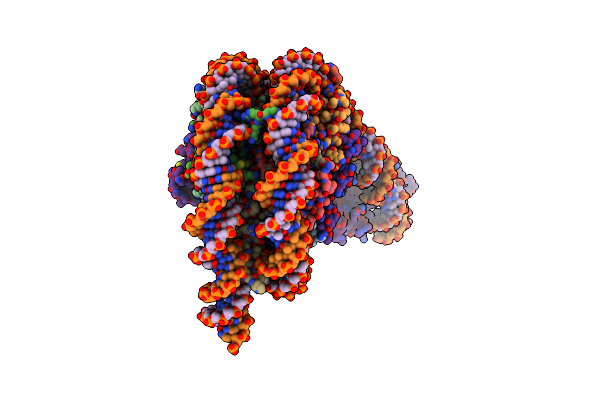 |
Composite Model Of A Chd1-Nucleosome Complex In The Nucleotide-Free State Derived From 2.3A And 2.7A Cryo-Em Maps
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: DNA BINDING PROTEIN |
 |
Structure Of Nf-Kb P52 Homodimer Bound To 13-Mer A/T-Centric P-Selectin Kb Dna Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-01-26 Classification: DNA BINDING PROTEIN-DNA/DNA |

