Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
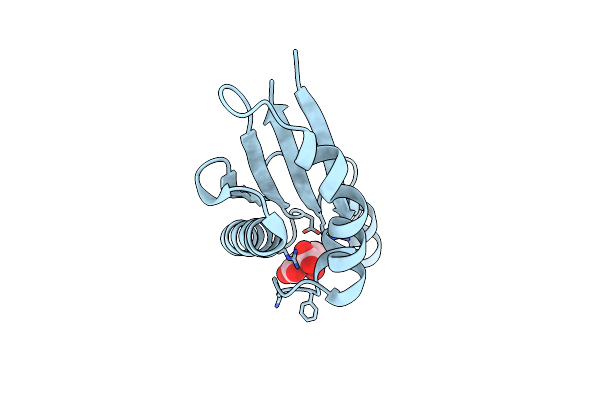
Crystal Structure Of (3R)-Hydroxyacyl-Acp Dehydratase Hadbd From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-27 Classification: LYASE |
 |
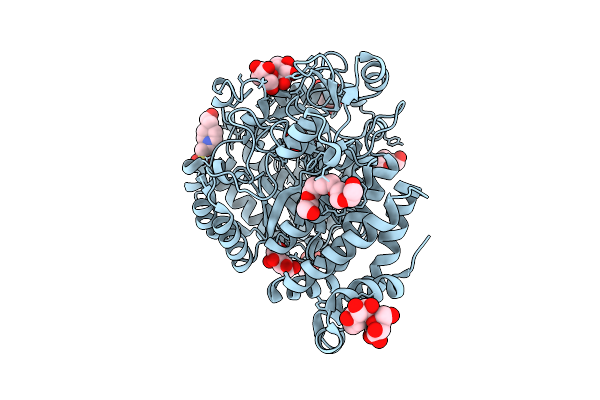
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-05-16 Classification: MEMBRANE PROTEIN Ligands: CIT |
 |
Crystal Structure Of Neisseria Polysaccharea Amylosucrase Mutant Derived From Neutral Genetic Drift-Based Engineering
Organism: Neisseria polysaccharea
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: TRS, GLC, EPE, PGE, FRU, 1PE |
 |
Crystal Structure Of Neisseria Polysaccharea Amylosucrase Mutant Efficient For The Synthesis Of Controlled Size Maltooligosaccharides
Organism: Neisseria polysaccharea
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-06-28 Classification: HYDROLASE Ligands: TRS, 1PE, PG4 |
 |
N-Terminally Truncated Dextransucrase Dsr-E From Leuconostoc Mesenteroides Nrrl B-1299 In Complex With D-Glucose
Organism: Leuconostoc mesenteroides subsp. mesenteroides
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-08-05 Classification: TRANSFERASE Ligands: CA, BGC, GLC, PEG |
 |
N-Terminally Truncated Dextransucrase Dsr-E From Leuconostoc Mesenteroides Nrrl B-1299 In Complex With Isomaltotriose
Organism: Leuconostoc mesenteroides
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: CA, NA, GLC, PEG |
 |
N-Terminally Truncated Dextransucrase Dsr-E From Leuconostoc Mesenteroides Nrrl B-1299 In Complex With Gluco-Oligosaccharides
Organism: Leuconostoc mesenteroides
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: CA, NA, GLC, GOL |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.60 Angstrom In Complex With N-Acetylglucosamine And Inorganic Phosphate
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, NDG, GOL, K, EDO |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.85 Angstrom From Unknown Human Gut Bacteria (Uhgb_Mp)
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, GOL, K, EDO, PGE |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.60 Angstrom In Complex With Beta-D-Mannopyranose And Inorganic Phosphate
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, BMA, K, EDO, PGE |
 |
Crystal Structure Of B-1,4-Mannopyranosyl-Chitobiose Phosphorylase At 1.76 Angstrom From Unknown Human Gut Bacteria (Uhgb_Mp) In Complex With N-Acetyl-D-Glucosamine, Beta-D-Mannopyranose And Inorganic Phosphate
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Release Date: 2015-05-27 Classification: TRANSFERASE Ligands: PO4, BMA, NDG, PGE, EDO, K, GOL |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Umabf62A From Ustilago Maidys
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, GOL, TRS, 1PE |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Umabf62A From Ustilago Maydis In Complex With L-Arabinofuranose
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, FUB, AHR, TRS |
 |
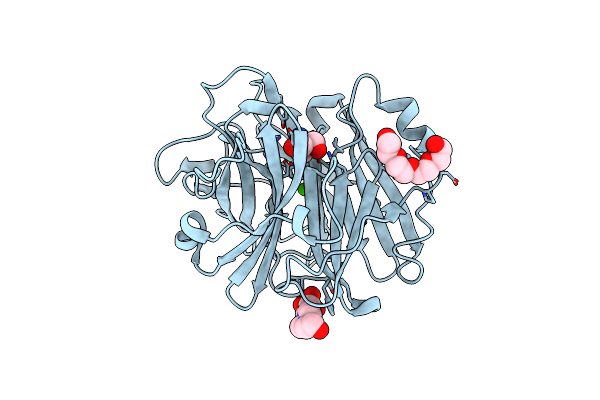
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina In Complex With Cellotriose
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, TRS, 1PE, EPE |
 |
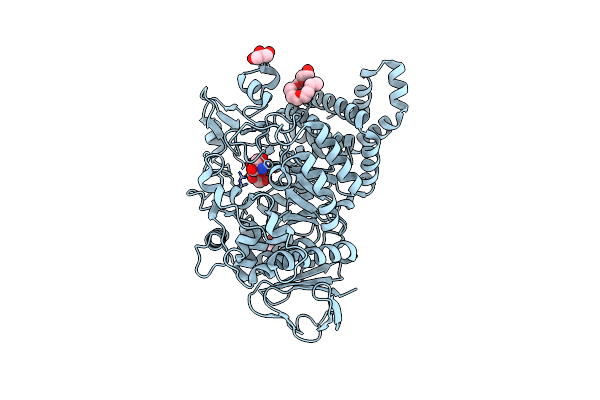
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, EPE, TRS, 1PE |
 |
Crystal Structure Of Amylosucrase Double Mutant A289P-F290C From Neisseria Polysaccharea
Organism: Neisseria polysaccharea
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-10-31 Classification: TRANSFERASE Ligands: TRS, GOL, P6G |
 |
Crystal Structure Of Amylosucrase Double Mutant A289P-F290I From Neisseria Polysaccharea.
Organism: Neisseria polysaccharea
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-10-31 Classification: TRANSFERASE Ligands: TRS, GOL, PGE, PG4 |
 |
Crystal Structure Of Amylosucrase Double Mutant A289P-F290L From Neisseria Polysaccharea
Organism: Neisseria polysaccharea
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-10-31 Classification: TRANSFERASE Ligands: TRS, GOL, 1PE |
 |
Crystal Structure Of Amylosucrase Inactive Double Mutant F290K-E328Q From Neisseria Polysaccharea In Complex With Sucrose.
Organism: Neisseria polysaccharea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-31 Classification: TRANSFERASE Ligands: CL, GOL |
 |
Crystal Structure Of Leuconostoc Mesenteroides Nrrl B-1299 N-Terminally Truncated Dextransucrase Dsr-E In Triclinic Form
Organism: Leuconostoc mesenteroides
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: CA, GOL |