Search Count: 76
 |
Organism: Synthetic construct, Metagenome
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
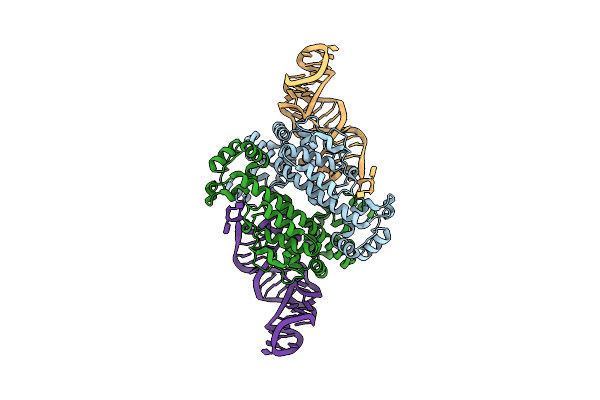 |
Organism: Uncultured prevotellaceae bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TOXIN/RNA |
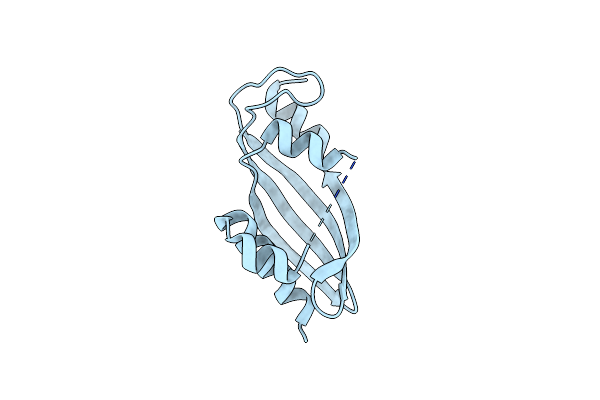 |
Crystal Structure Of Dyna1, A Putative Monoxygenase From Mivromonospora Chersina.
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Organism: Selenomonas sp., Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Cib_12 Beta-Galactosidase From Cuniculiplasma Divulgatum
Organism: Cuniculiplasma divulgatum
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Cib_13 Beta-Galactosidase From Cuniculiplasma Divulgatum
Organism: Cuniculiplasma divulgatum
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: GOL, TRS |
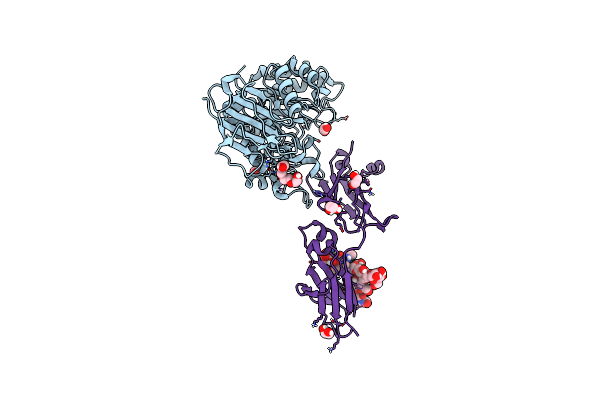 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-10-18 Classification: TRANSPORT PROTEIN Ligands: EDO, PG4 |
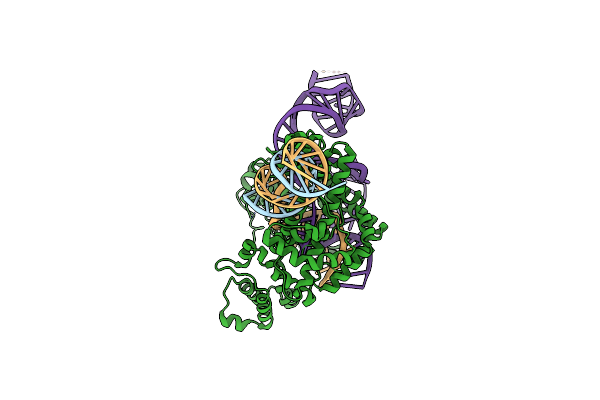 |
Structure Of The Spizellomyces Punctatus Fanzor (Spufz) In Complex With Omega Rna And Target Dna
Organism: Methanosarcina mazei, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
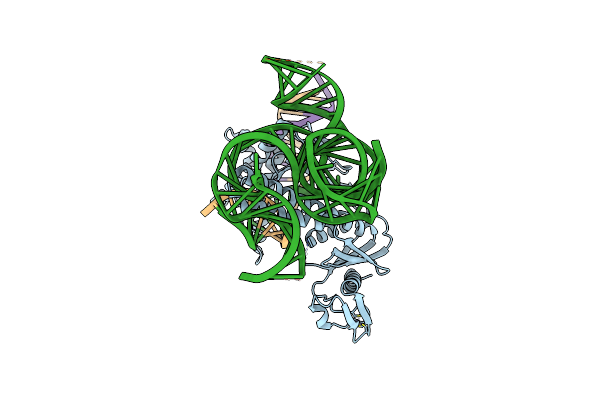 |
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: RNA BINDING PROTEIN Ligands: ZN |
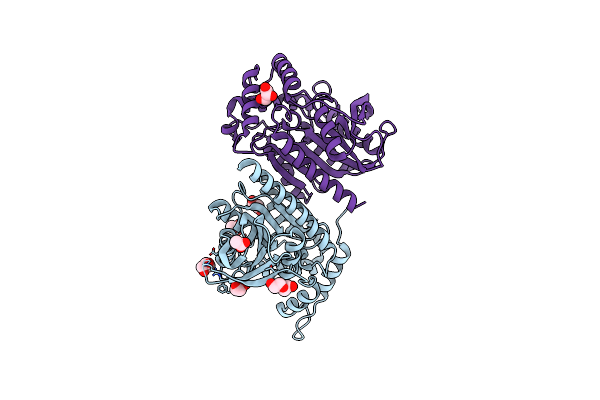 |
Structure Of Ester-Hydrolase Eh0 From The Metagenome Of Sorghum Bicolor Rhizosphere From The Henfaes Research Centre (Gwynedd, Wales)
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: ACT, CL, GOL, PEG |
 |
Organism: Metagenome, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, LDA |
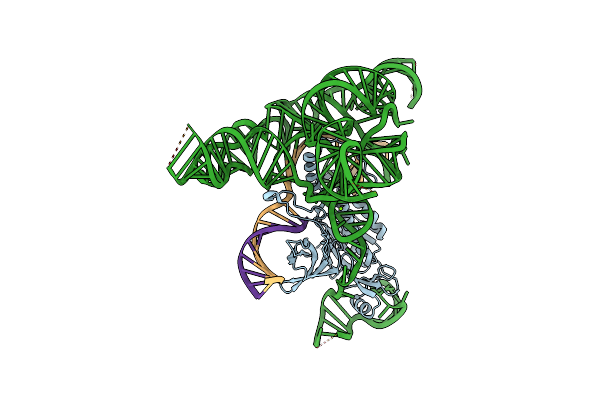 |
Structure Of Desulfovirgula Thermocuniculi Isrb (Dtisrb) In Complex With Omega Rna And Target Dna
Organism: Saccharomyces cerevisiae s288c, Desulfovirgula thermocuniculi, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-10-19 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG |
 |
Organism: Planctomycetes bacterium, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.38 Å Release Date: 2022-09-07 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Planctomycetes bacterium, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-08-24 Classification: RNA BINDING PROTEIN/RNA Ligands: CL, EDO, BR, MG |
 |
 |
 |
Pharmacological Characterisation And Nmr Structure Of The Novel Mu-Conotoxin Sxiiic, A Potent Irreversible Nav Channel Inhibitor
|
 |
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2020-07-08 Classification: CHAPERONE Ligands: GOL, SO4 |
 |
Structure Of The Pore C-Terminal Domain, A Protein Of T9Ss From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis jcvi sc001
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-06-17 Classification: PROTEIN BINDING Ligands: MLD, NA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-11-06 Classification: cytosolic protein, transferase Ligands: MG, N7Y |

