Search Count: 33
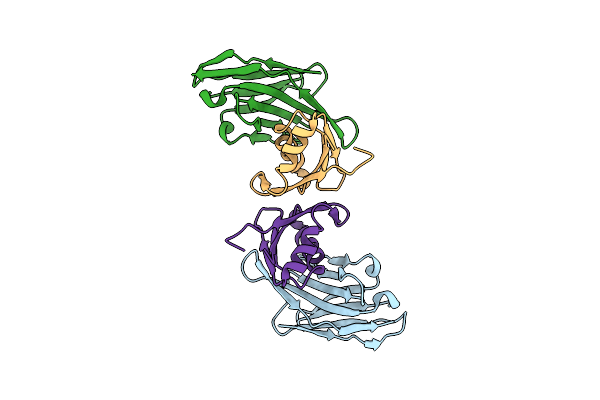 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-02-28 Classification: TRANSPORT PROTEIN |
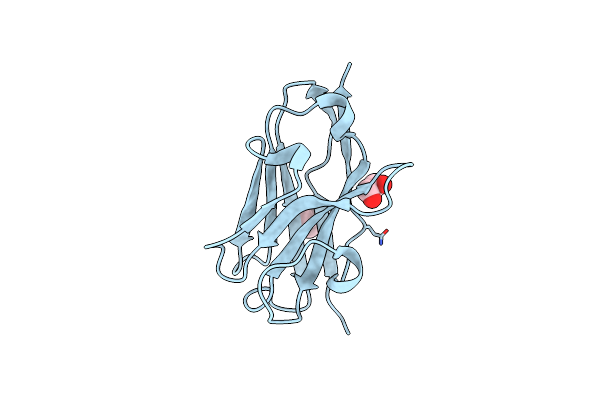 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: EDO, PEG |
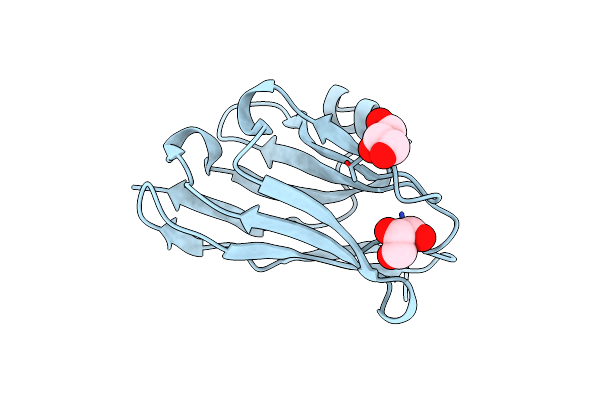 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: GOL |
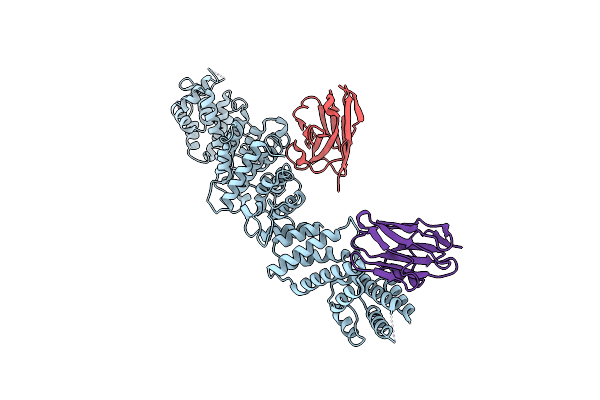 |
Organism: Xenopus laevis, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-11-08 Classification: NUCLEAR PROTEIN |
 |
Crystal Structure Of Nucleoporin-98 Nanobody Ms98-6 Complex Solved At 2.2A Resolution
Organism: Xenopus tropicalis, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-21 Classification: NUCLEAR PROTEIN Ligands: TRS, NA |
 |
Organism: Vicugna pacos, Xenopus tropicalis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-07 Classification: NUCLEAR PROTEIN |
 |
High Resolution Crystal Structure Of C-Terminal Domain (Residues 715-866) Of Nucleoporin-98
Organism: Xenopus tropicalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-04-07 Classification: NUCLEAR PROTEIN Ligands: MPD, NA, CL |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2019-05-01 Classification: PROTEIN TRANSPORT Ligands: GTP, MG |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:4.53 Å Release Date: 2019-05-01 Classification: PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2019-05-01 Classification: PROTEIN TRANSPORT Ligands: GTP, MG |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-06-22 Classification: PROTEIN TRANSPORT Ligands: GTP, MG |
 |
Organism: Vicugna pacos, Xenopus tropicalis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-12-16 Classification: TRANSPORT PROTEIN |
 |
Organism: Camelus dromedarius, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-08-26 Classification: TRANSPORT PROTEIN |
 |
Organism: Xenopus laevis, Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-08-26 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of A 70S Ribosome-Trna Complex Reveals Functional Interactions And Rearrangements.
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.83 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, ZN |
 |
Crystal Structure Of A Translation Termination Complex Formed With Release Factor Rf2.
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, ZN |
 |
Crystal Structure Of Release Factor Rf3 Trapped In The Gtp State On A Rotated Conformation Of The Ribosome.
Organism: Escherichia coli, Streptomyces
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-07-09 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, GNP |
 |
Crystal Structure Of Release Factor Rf3 Trapped In The Gtp State On A Rotated Conformation Of The Ribosome (Without Viomycin)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: GNP, MG |

