Search Count: 191
 |
Organism: Francisella tularensis subsp. tularensis schu s4
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: ZN, PEG |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: GOL, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: GOL, FLC |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FLC |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: GOL, IPA, CIT, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: GOL |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: PEG, P6G, GOL, MG, CL, SO4 |
 |
Structure Of Caulobacter Crescentus Suppressor Of Copper Sensitivity Protein C
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: PEG, TCH |
 |
Crystal Structure Of Vibrio Cholerae Dsba In Complex With Bile Salt Taurocholate
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: GOL, TCH |
 |
Crystal Structure Of E.Coli Dsba In Complex With 2-(2,6-Bis(3-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VCY, CU |
 |
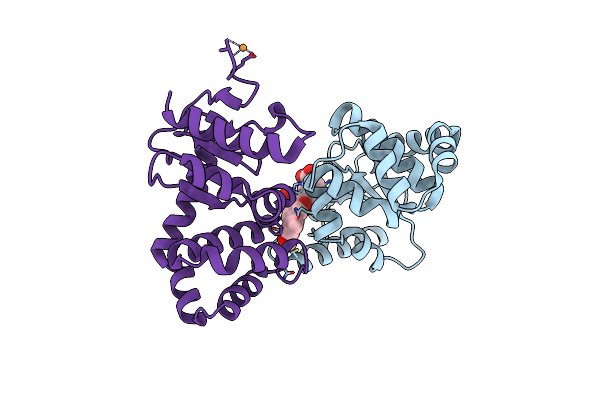
Crystal Structure Of E.Coli Dsba In Complex With 2-(6-(3-Methoxyphenyl)-2-(4-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VE7, CU |
 |
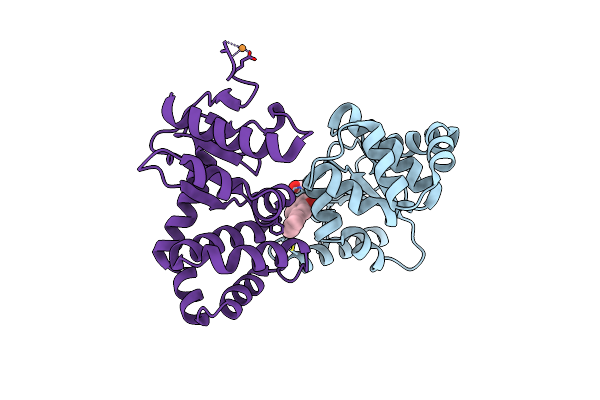
Crystal Structure Of E.Coli Dsba In Complex With 3-(3-(Carboxymethyl)-6-(3-Methoxyphenyl)Benzofuran-2-Yl)Benzoic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: VED, CU |
 |
Crystal Structure Of Ecdsba In A Complex With 2-(6-Phenylbenzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XPV, CU |
 |
Crystal Structure Of Ecdsba In A Complex With 2-(6-(3-Methoxyphenyl)Benzofuran-3-Yl)Acetic Acid
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XQ1, CU |
 |
Crystal Structure Of Ecdsba In A Complex With Methyl 2-(6-Bromo-2-Phenylbenzofuran-3-Yl)Acetate
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-08-11 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: Y1G, CU |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: MG, EDO, PGE |
 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product F1 (Methylpiperazinone 6)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O7P, CU, EDO |

