Search Count: 19
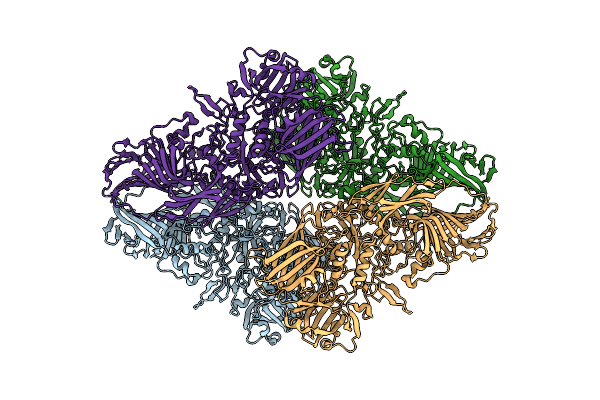 |
Cryo-Em Structure Of Beta-Galactosidase At 3.3 A Resolution Plunged 5 Ms After Mixing With Apoferritin
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: HYDROLASE |
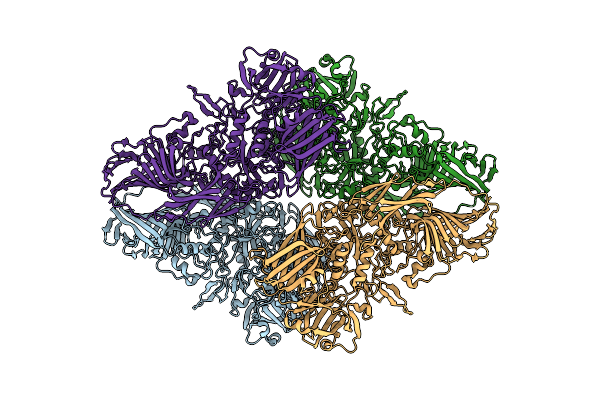 |
Cryo-Em Structure Of Beta-Galactosidase At 2.9 A Resolution Plunged 205 Ms After Mixing With Apoferritin
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: HYDROLASE |
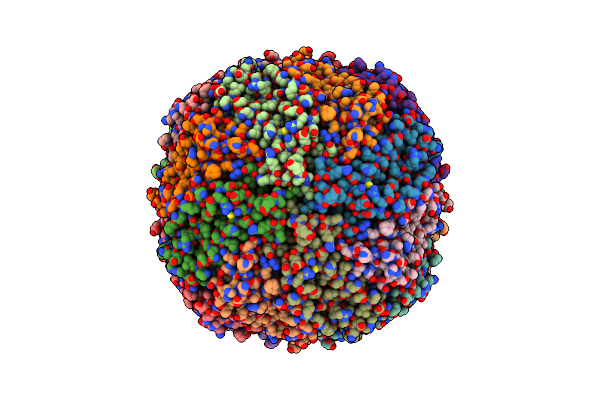 |
Cryo-Em Structure Of Mouse Heavy-Chain Apoferritin At 2.1 A Plunged 5Ms After Mixing With B-Galactosidase
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: METAL BINDING PROTEIN Ligands: FE |
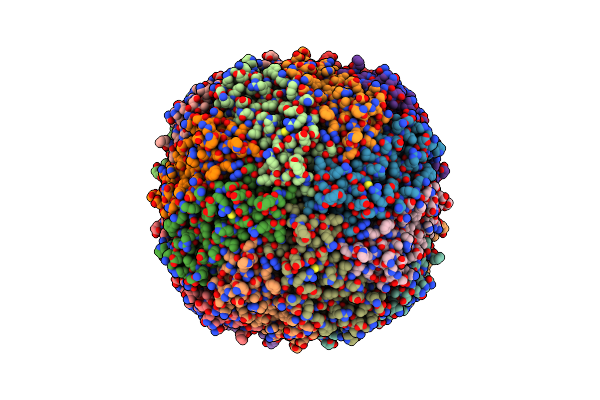 |
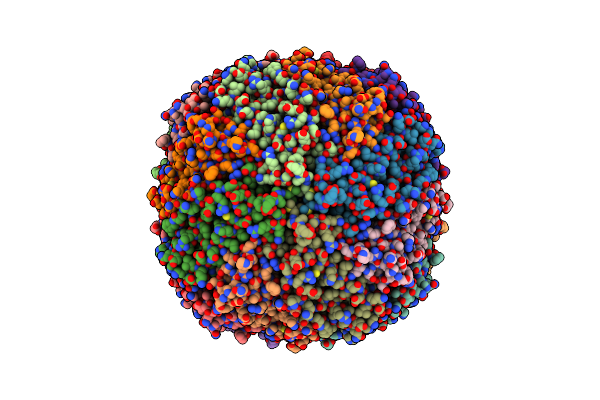
Cryo-Em Structure Of Mouse Heavy-Chain Apoferritin At 2.7 A Plunged 35Ms After Mixing With B-Galactosidase
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
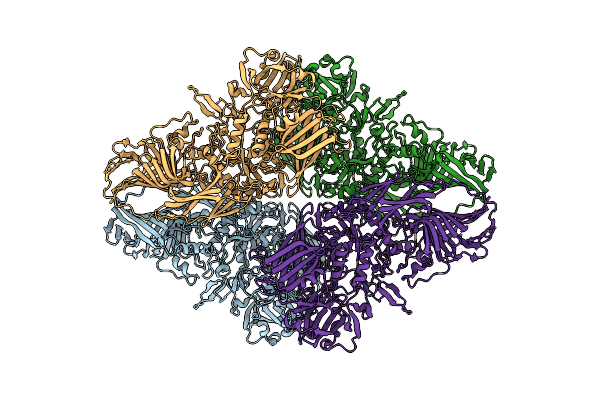
Cryo-Em Structure Of Mouse Heavy-Chain Apoferritin At 2.2 A Plunged 205Ms After Mixing With B-Galactosidase
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
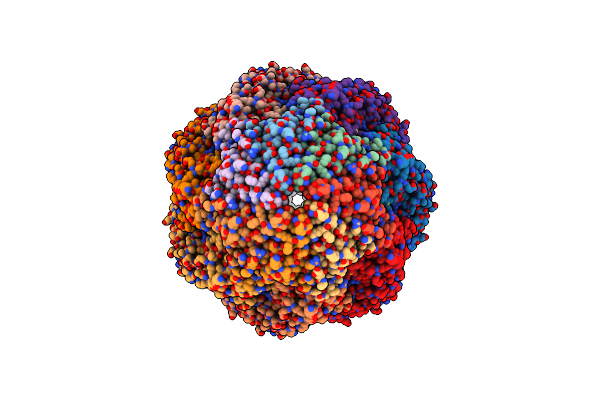
Cryo-Em Structure Of Beta-Galactosidase At 3.2 A Resolution Plunged 35 Ms After Mixing With Apoferritin
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: HYDROLASE |
 |
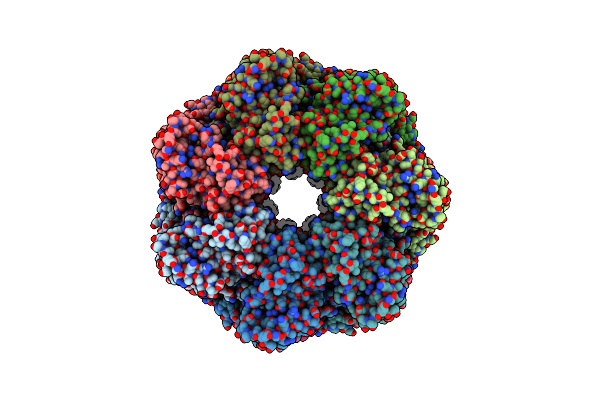
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, K, ATP |
 |
Structure Of Groel-Atp Complex Plunge Frozen 200 Ms After Reaction Initiation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, ATP, K |
 |
Structure Of Groel-Nucleotide Complex In Adp-Like Conformation Plunged 13 Ms After Mixing With Atp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: ATP |
 |
Structure Of The Groel-Atp Complex Plunge-Frozen 50 Ms After Mixing With Atp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, ATP, K |
 |
Structure Of The Groel(Atp7/Adp7) Complex Plunged 13 Ms After Mixing With Atp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: ATP |
 |
Structure Of Groel-Nucleotide Complex In Adp-Like Conformation Plunged 50 Ms After Mixing With Atp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: ATP |
 |
Structure Of The Groel(Atp7/Adp7) Complex Plunged 50 Ms After Mixing With Atp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: ATP |
 |
Structure Of The Groel-Atp Complex Plunge-Frozen 13 Ms After Mixing With Atp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, ATP |
 |
Structure Of Groel:Groes-Atp Complex Plunge Frozen 200 Ms After Reaction Initiation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, K, ATP |
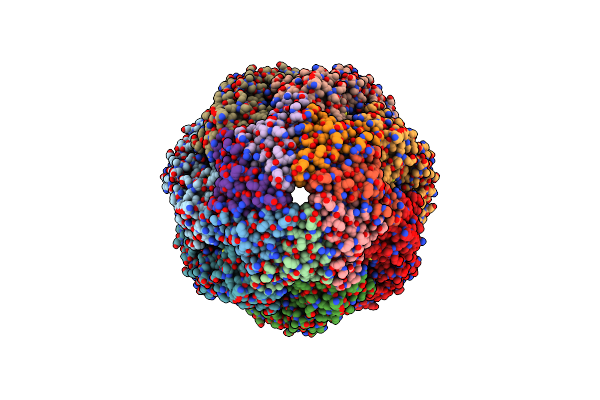 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, K, ATP |
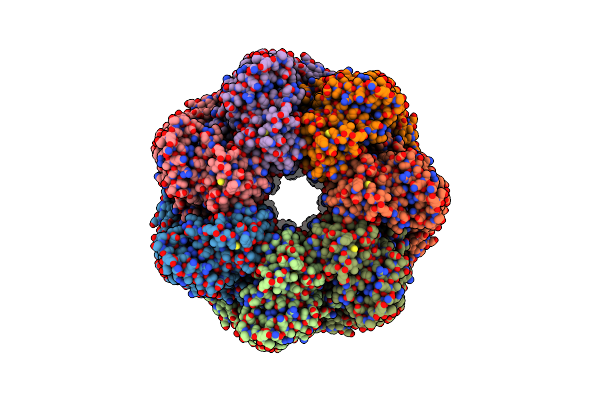 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, ATP, K |
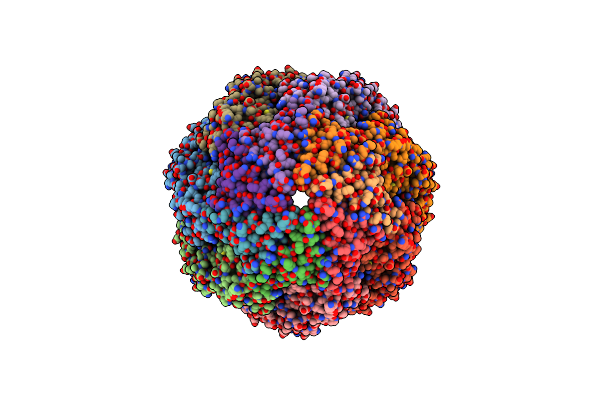 |
Structure Of Groel:Groes Complex Exhibiting Adp-Conformation In Trans Ring Obtained Under The Continuous Turnover Conditions
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, ATP |
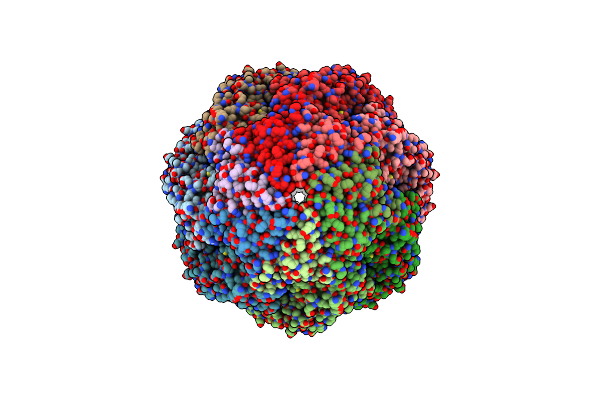 |
Structure Of Groel:Groes-Atp Complex Plunge Frozen 200 Ms After Reaction Initiation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CHAPERONE Ligands: MG, K, ATP |

