Search Count: 20
 |
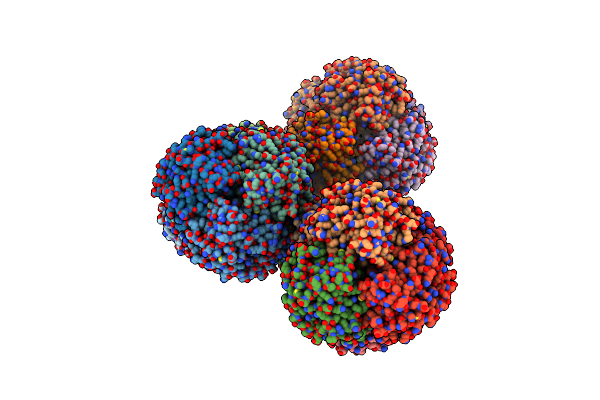
Cryo-Em Structure Of Burkholderia Cenocepacia Orotate Phosphoribosyltransferase
Organism: Burkholderia cenocepacia j2315
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: TRANSFERASE Ligands: ACT |
 |
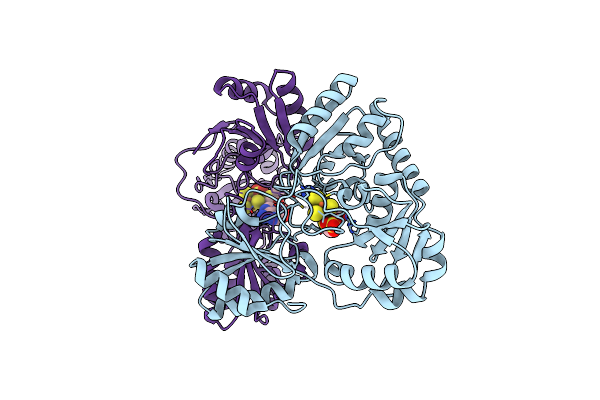
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: MN |
 |
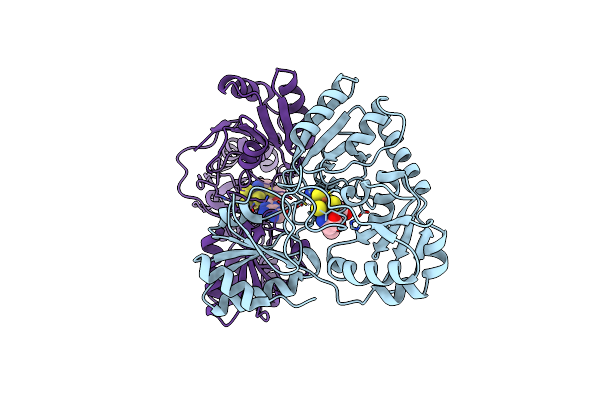
Crystal Structure Of Pyrococcus Horikoshii Dph2 With 4Fe-4S Cluster And Mta
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-04-11 Classification: BIOSYNTHETIC PROTEIN Ligands: MTA, SO4, SF4 |
 |
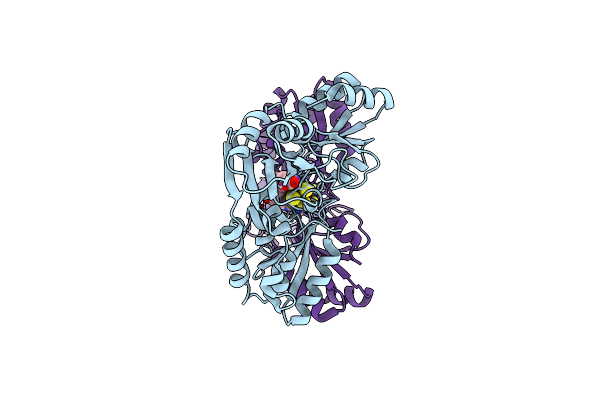
Crystal Structure Of Pyrococcus Horikoshii Dph2 With 4Fe-4S Cluster And Sam
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-04-11 Classification: BIOSYNTHETIC PROTEIN Ligands: SAM, SF4 |
 |
Crystal Structure Of Candidatus Methanoperedens Nitroreducens Dph2 With 4Fe-4S Cluster And Sam/Cleaved Sam
Organism: Candidatus methanoperedens nitroreducens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-04-11 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, SAM, ABA, MTA |
 |
Crystal Structure Of Candidatus Methanoperedens Nitroreducens Dph2 With 4Fe-4S Cluster And Sam
Organism: Candidatus methanoperedens nitroreducens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2018-04-11 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, SAM |
 |
Crystal Structure Of Candidatus Methanoperedens Nitroreducens Dph2 With 4Fe-4S Cluster And Sah
Organism: Candidatus methanoperedens nitroreducens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-04-11 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, SAH |
 |
Crystal Structure Of A Short Chain Dehydrogenase From Burkholderia Cenocepacia J2315 In Complex With Nadp.
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Release Date: 2017-01-18 Classification: OXIDOREDUCTASE Ligands: NAP, BEZ, PO4 |
 |
Structure Of A Putative Phosphomethylpyrimidine Kinase From Acinetobacter Baumannii In Non-Covalent Complex With Pyridoxal Phosphate
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Brucella ovis intabari-2002-82-58
Method: X-RAY DIFFRACTION Release Date: 2016-02-10 Classification: OXIDOREDUCTASE Ligands: IMD, EDO, NAD |
 |
Organism: Brucella ovis (strain atcc 25840 / 63/290 / nctc 10512)
Method: X-RAY DIFFRACTION Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: ACT, EDO |
 |
Structure Of A Putative Phosphomethylpyrimidine Kinase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii is-123
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-04-01 Classification: TRANSFERASE Ligands: EDO |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-07-14 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, SO4 |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2010-06-23 Classification: BIOSYNTHETIC PROTEIN |
 |
The Structural Basis For Recognition Of The Preq0 Metabolite By An Unusually Small Riboswitch Aptamer Domain
Method: X-RAY DIFFRACTION
Resolution:2.75 Å Release Date: 2009-03-03 Classification: RNA Ligands: PQ0, SO4 |
 |
A 3'-Oh, 2',5'-Phosphodiester Substitution In The Hairpin Ribozyme Active Site Reveals Similarities With Protein Ribonucleases
Method: X-RAY DIFFRACTION
Resolution:2.80 Å Release Date: 2008-05-20 Classification: RNA Ligands: NCO |
 |
A Minimal, 'Hinged' Hairpin Ribozyme Construct Solved With Mimics Of The Product Strands At 2.25 Angstroms Resolution
Method: X-RAY DIFFRACTION
Resolution:2.25 Å Release Date: 2007-05-22 Classification: RNA Ligands: SO4, NA, NCO |
 |
Vanadate At The Active Site Of A Small Ribozyme Suggests A Role For Water In Transition-State Stabilization
Method: X-RAY DIFFRACTION
Resolution:2.05 Å Release Date: 2007-05-22 Classification: RNA Ligands: SO4, NCO |
 |
The Novel Use Of A 2',5'-Phosphodiester Linkage As A Reaction Intermediate At The Active Site Of A Small Ribozyme
Method: X-RAY DIFFRACTION
Resolution:2.35 Å Release Date: 2007-05-22 Classification: RNA Ligands: SO4, NCO |
 |
The Crystal Structure Of Cytidine Deaminase Cdd1, An Orphan C To U Editase From Yeast
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-25 Classification: HYDROLASE Ligands: ZN |

