Search Count: 27
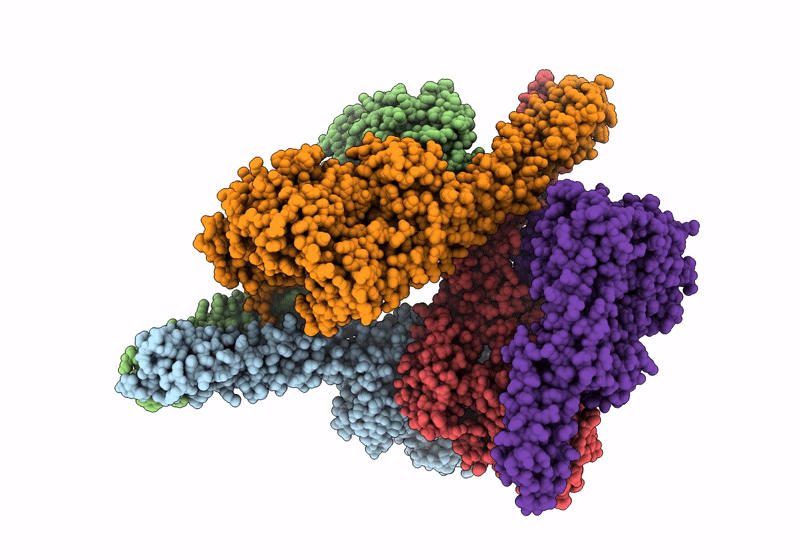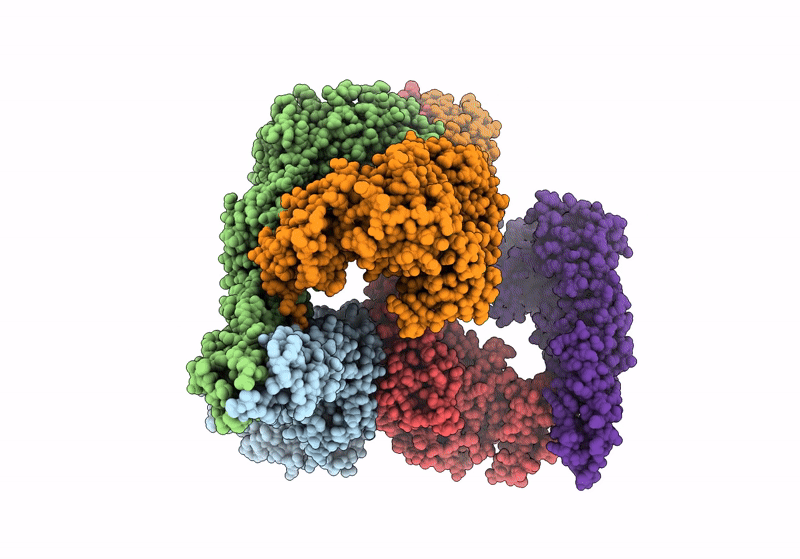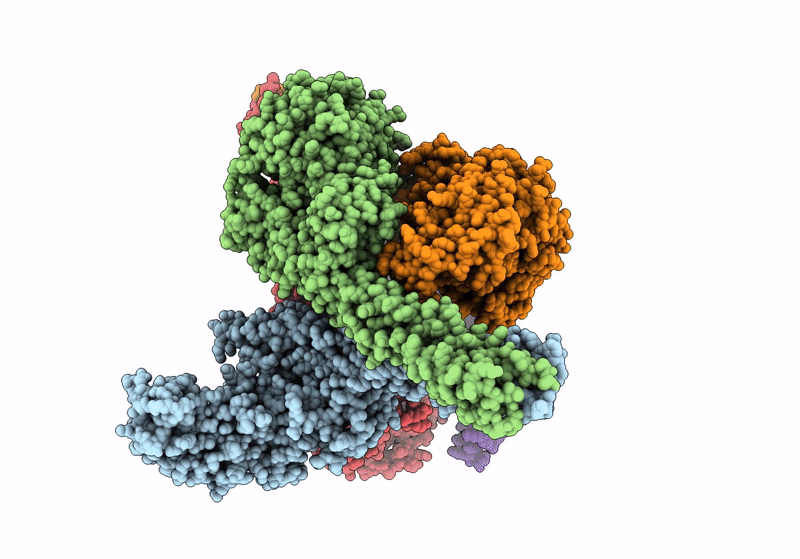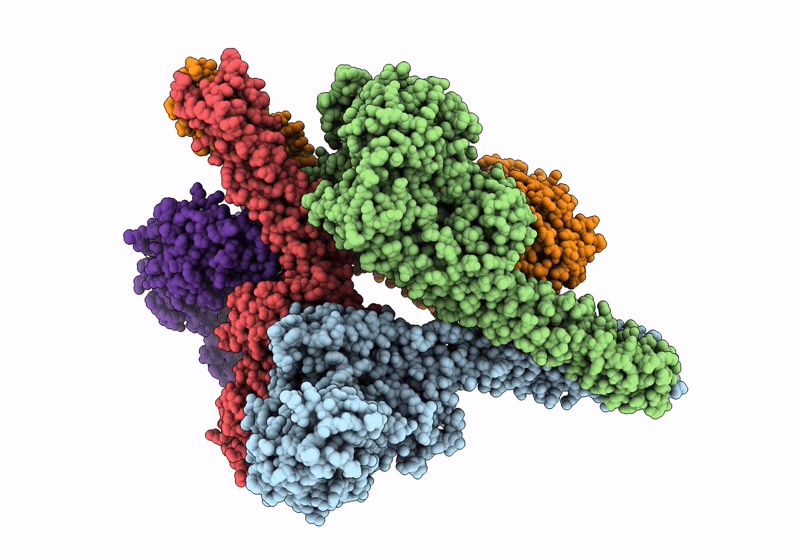 |
Crystal Structure Of The Constitutively Active S117E/S181E Mutant Of Human Ikk2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.16 Å Release Date: 2024-04-10 Classification: TRANSFERASE |
 |
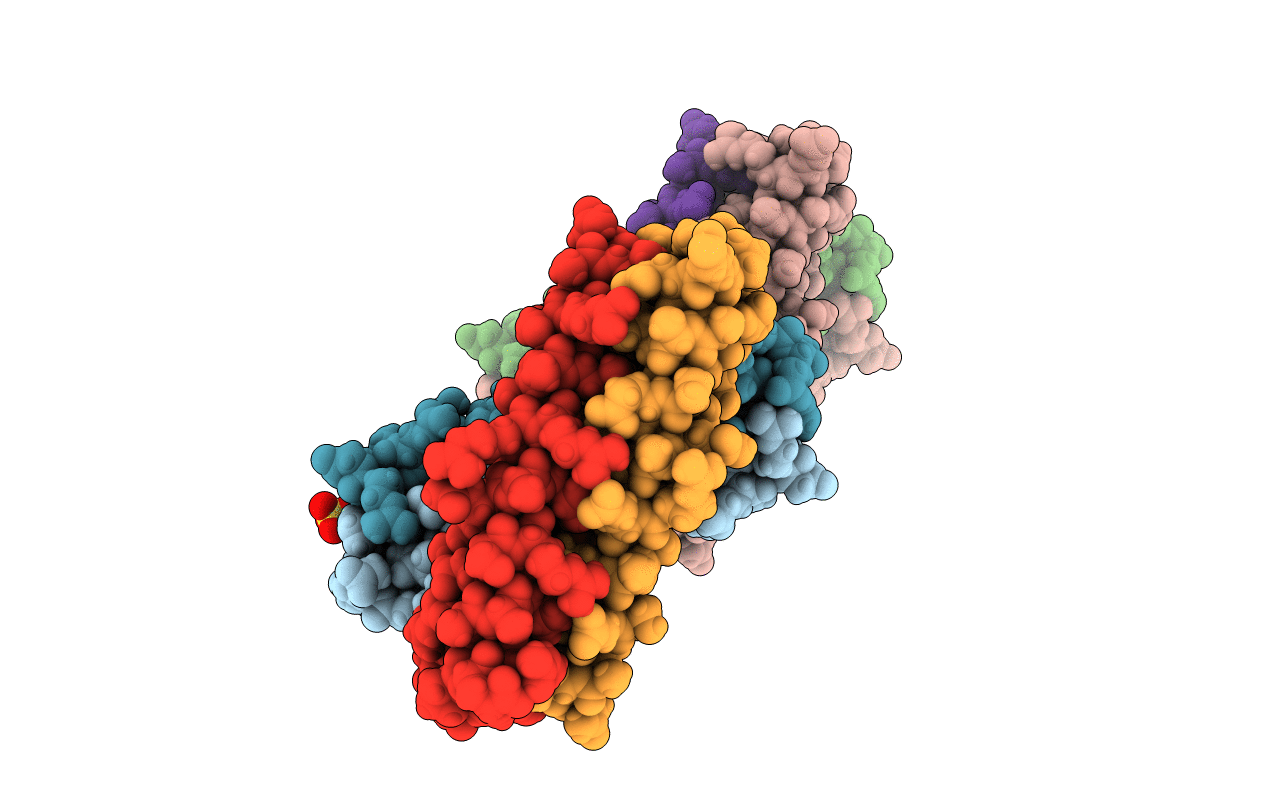
Het-N2 - De Novo Designed Three-Helix Heterodimer With Cysteine At The N2 Position Of The Alpha-Helix
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2021-03-17 Classification: DE NOVO PROTEIN Ligands: CD, NA |
 |
Het-Ncap - De Novo Designed Three-Helix Heterodimer With Cysteine At The Ncap Position Of The Alpha-Helix
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-03-17 Classification: DE NOVO PROTEIN Ligands: SO4, ZN |
 |
Het-N2-So3- - De Novo Designed Three-Helix Heterodimer With Cysteine S-Sulfate At The N2 Position Of The Alpha-Helix
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-03-17 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
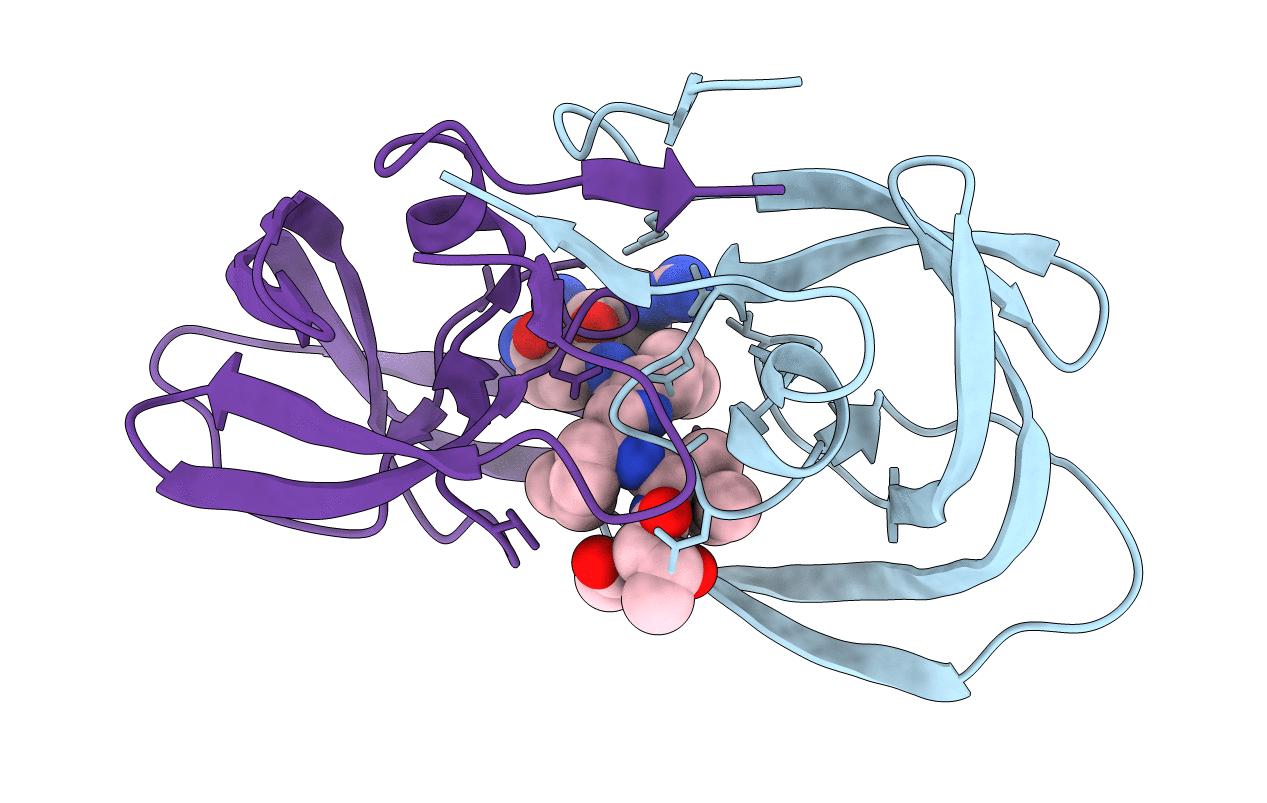
Crystal Structure Of Complex Between Nuclear Coactivator Binding Domain Of Cbp And [1040-1086]Actr Containing Alpha-Methylated Leu1055 And Leu1076
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-09-30 Classification: PROTEIN BINDING Ligands: EDO, ZN |
 |
Organism: Crambe hispanica subsp. abyssinica
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2012-02-08 Classification: UNKNOWN FUNCTION |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Ile50,O-Ile50']Hiv-1 Protease Complexed With Mvt-101
Method: X-RAY DIFFRACTION
Resolution:1.50 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Ile50,O-Ile50']Hiv-1 Protease Complexed With Kvs-1 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.90 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: SO4, KVS |
 |
X-Ray Structure Of Ester Chemical Analogue 'Covalent Dimer' [Ile50,O-Ile50']Hiv-1 Protease Complexed With Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.61 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC, SO4 |
 |
X-Ray Structure Of Ester Chemical Analogue 'Covalent Dimer' [Ile50,O-Ile50']Hiv-1 Protease Complexed With Kvs-1 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.80 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: KVS, SO4 |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Gly51,O-Gly51']Hiv-1 Protease Complexed With Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC, SO4 |
 |
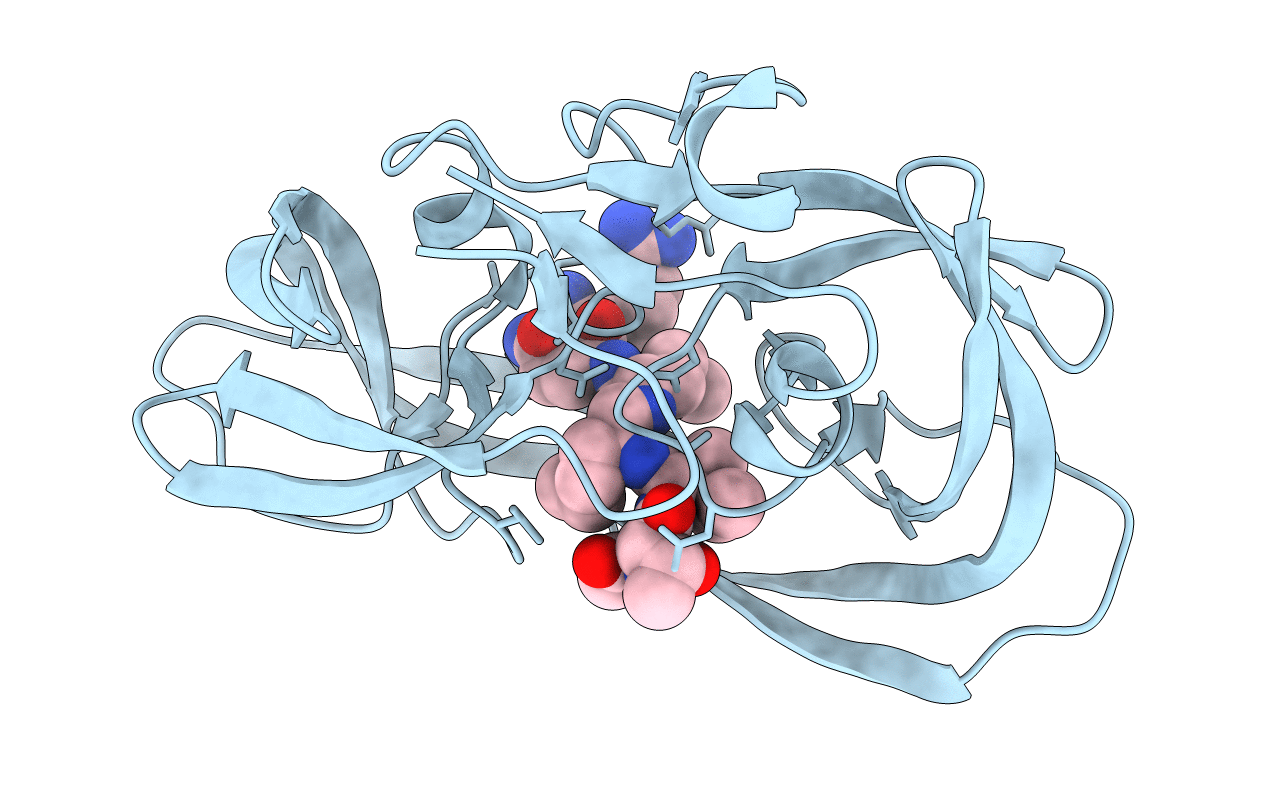
Crystal Structure Of A Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [L-Ala51,D-Ala51'] Hiv-1 Protease Molecule Complexed With Jg-365 Inhibitor
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-10 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
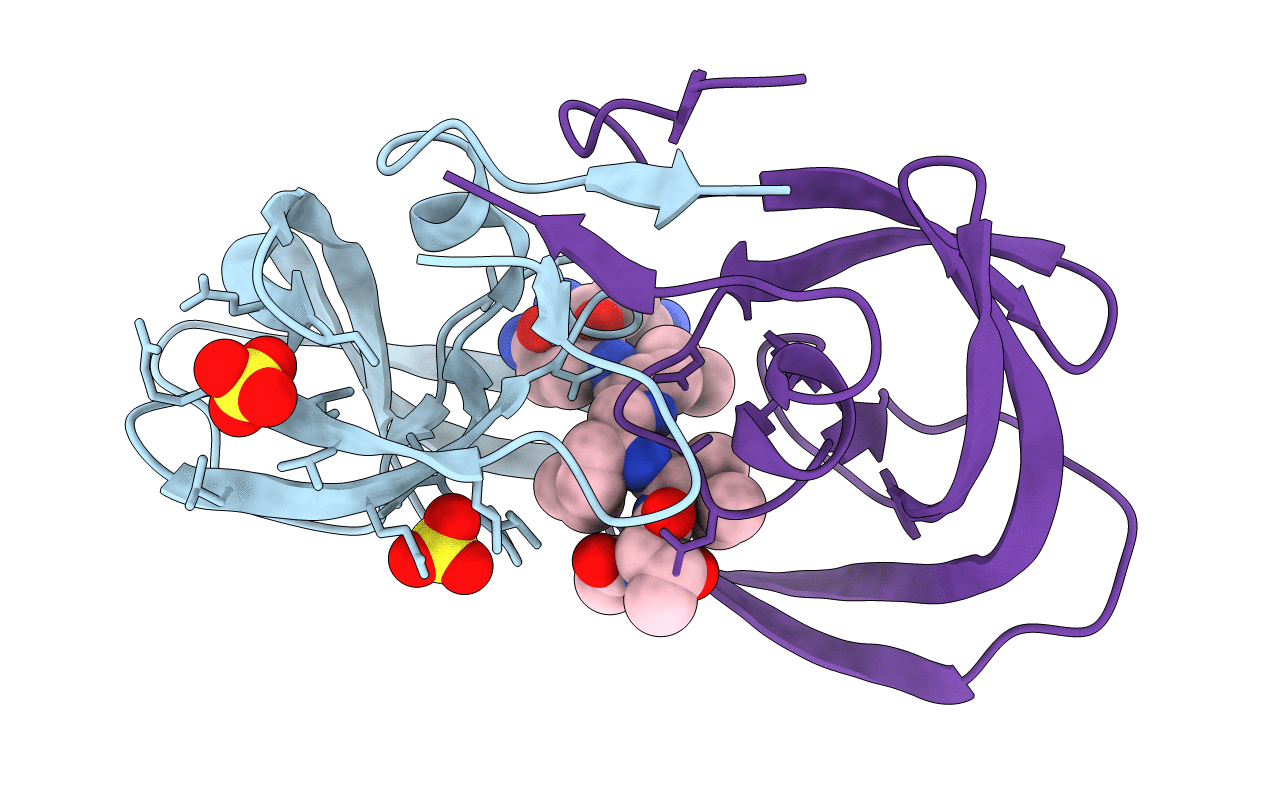
Crystal Structure Of Chemically Synthesized 'Covalent Dimer' [Gly51/D-Ala51']Hiv-1 Protease
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2011-07-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Crystal Structure Of A Chemically Synthesized [D25N]Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-07-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
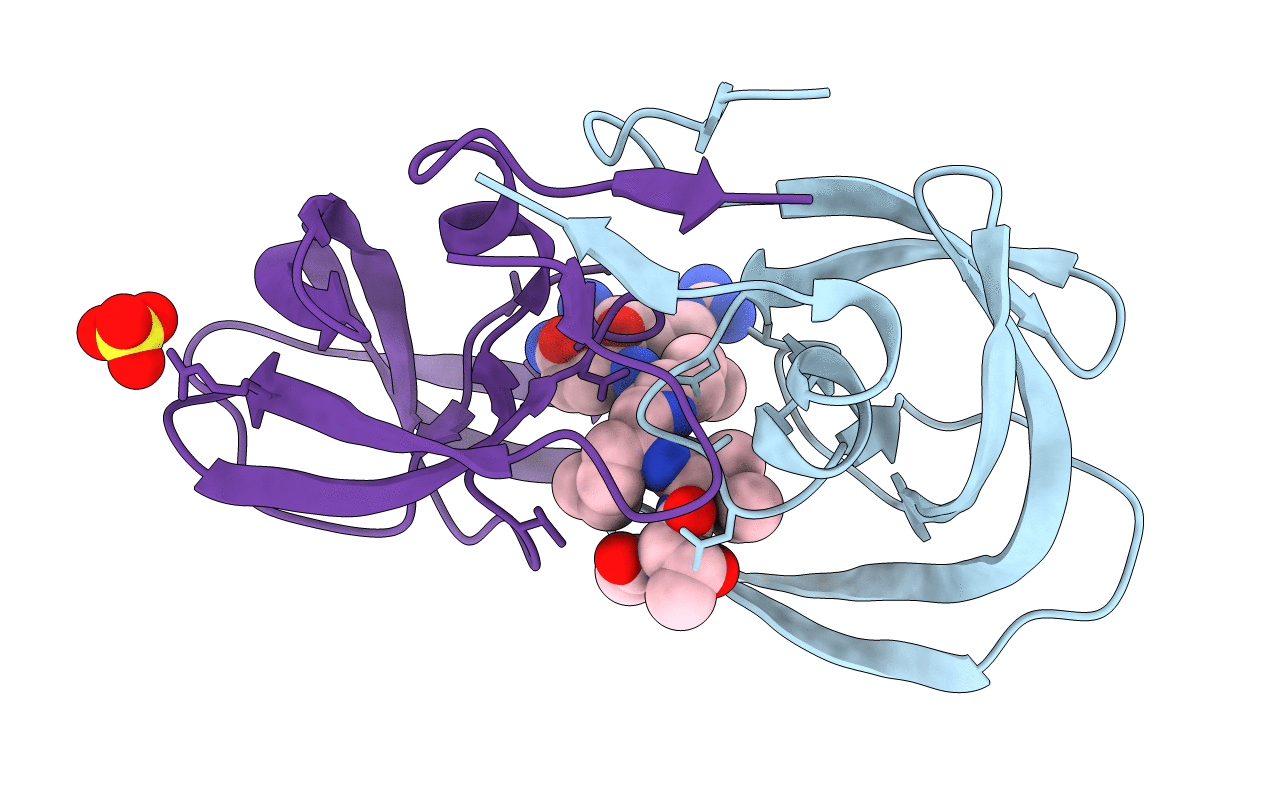
Crystal Structure Of Chemically Synthesized Hiv-1 Protease With Reduced Isostere Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
Crystal Structure Of [L-Ala51/51']Hiv-1 Protease With Reduced Isostere Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
Crystal Structure Of A Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [Gly51;Aib51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Crystal Structure Of A Chemically Synthesized [Allo-Ile50/50']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.50 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
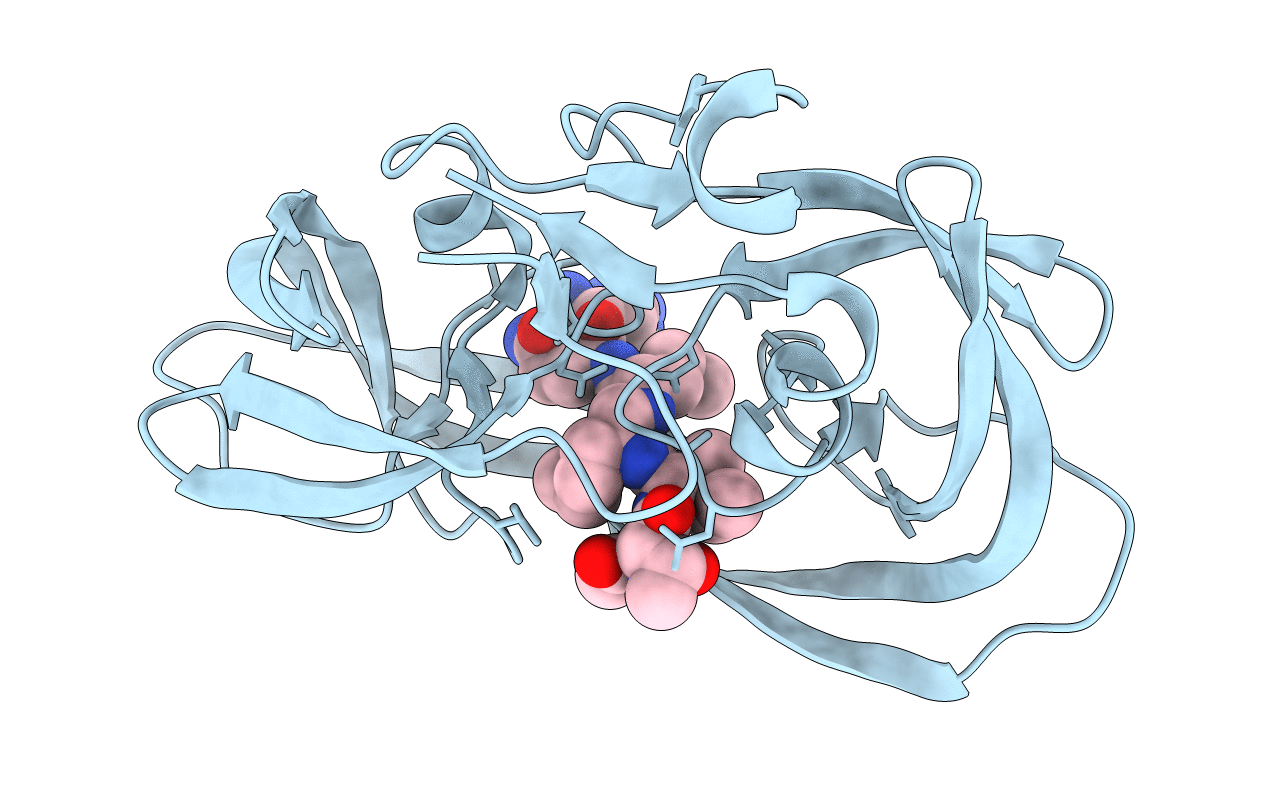
Crystal Structure Of A Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [Gly51;Aib51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor At 1.6 A Resolution
Method: X-RAY DIFFRACTION
Resolution:1.61 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
Crystal Structure Of Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [L-Ala;Gly51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor At 1.4 A Resolution
Method: X-RAY DIFFRACTION
Resolution:1.40 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |

