Search Count: 33
 |
Organism: Mayaro virus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-03-19 Classification: VIRAL PROTEIN Ligands: EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-31 Classification: Viral protein, HYDROLASE Ligands: GOL, PO4, ZN, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-05-17 Classification: Viral protein, HYDROLASE Ligands: PO4, GOL, ACT, ZN, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-06-22 Classification: RNA BINDING PROTEIN, Viral protein Ligands: GOL, ACT |
 |
Crystal Structure Of Sars-Cov-2 Nucleocapsid Protein C-Terminal Domain Complexed With Chicoric Acid
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-06-22 Classification: RNA BINDING PROTEIN/Viral Protein Ligands: GKP, PEG, GOL, NA, CL |
 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, GOL, PEG |
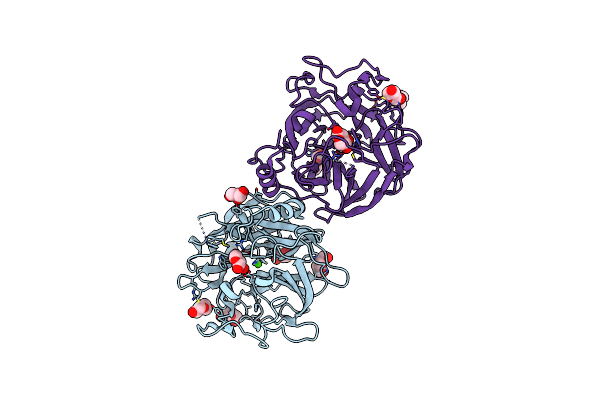 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
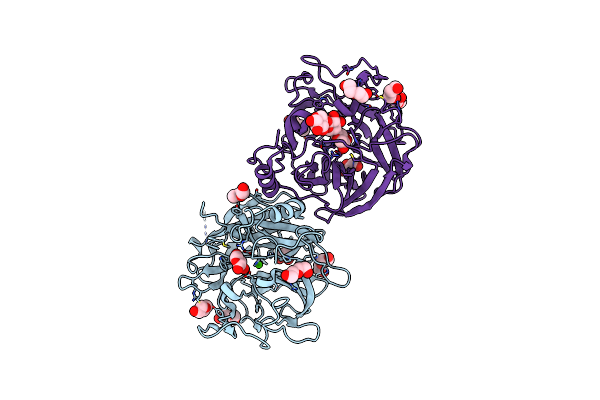 |
Crystal Structure Of The Gh43_1 Enzyme From Xanthomonas Citri Complexed With Xylotriose
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA, XYP, PEG, GOL |
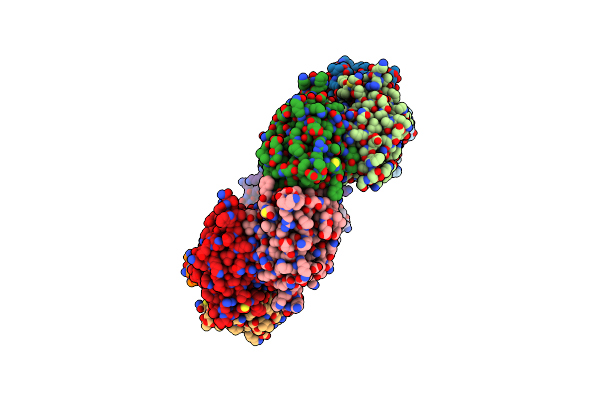 |
Yeast Thiol Specific Antoxidant 2 With C171S Mutation And Catalytic Cysteine Alkylated With Iodoacetamide
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-07-15 Classification: OXIDOREDUCTASE |
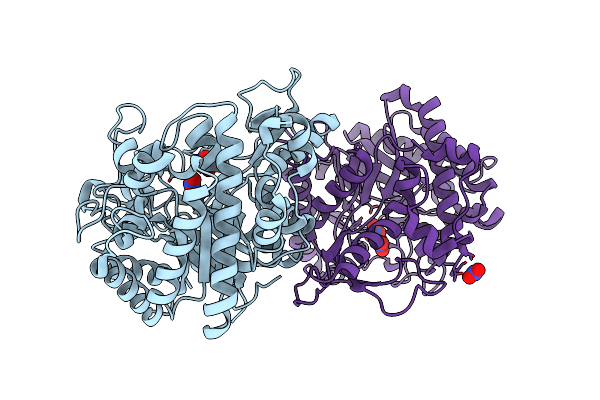 |
Crystal Structure Of The Double Mutant L167W / P172L Of The Beta-Glucosidase From Trichoderma Harzianum
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: NO3 |
 |
Crystal Structure Of The Gh51 Arabinofuranosidase From Xanthomonas Axonopodis Pv. Citri
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of A Gh1 Beta-Glucosidase Retrieved From Microbial Metagenome Of Poraque Amazon Lake
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Crystal Structure Of The Adenosine Kinase From Mus Musculus In Complex With Adenosine And Adenosine-Diphosphate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-07 Classification: TRANSFERASE Ligands: ADN, ADP, K, CL, MG, PG4, PO4 |
 |
High-Resolution Structure Of The Adenosine Kinase From Mus Musculus In Complex With Adenosine
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-06-07 Classification: TRANSFERASE Ligands: ADN, SO4, CL, PG4, ACT |
 |
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of The Gh1 Beta-Glucosidase From Exiguobacterium Antarcticum B7 In Space Group P21
Organism: Exiguobacterium antarcticum (strain b7)
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2016-04-13 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Gh1 Beta-Glucosidase From Exiguobacterium Antarcticum B7 In Space Group C2221
Organism: Exiguobacterium antarcticum (strain b7)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-04-13 Classification: HYDROLASE Ligands: CXS, SO4, GOL |
 |
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-02-05 Classification: HYDROLASE Ligands: TRS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-10-09 Classification: PROTEIN TRANSPORT Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2013-10-09 Classification: PROTEIN TRANSPORT Ligands: NO3 |

