Search Count: 84
 |
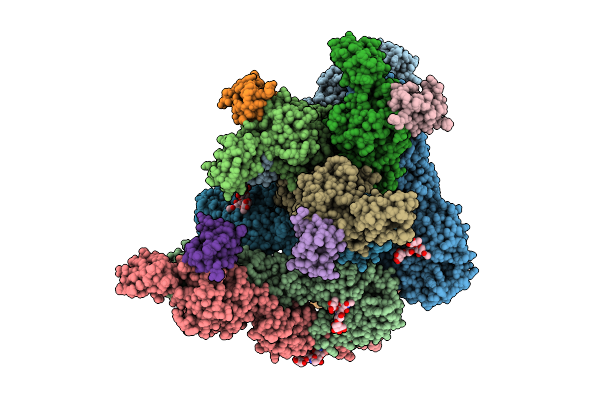
The 1:1 Cryo-Em Structure Of Bap1/Asxl1-K351Ub In Complex With H2Ak119Ub Nucleosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN/DNA |
 |
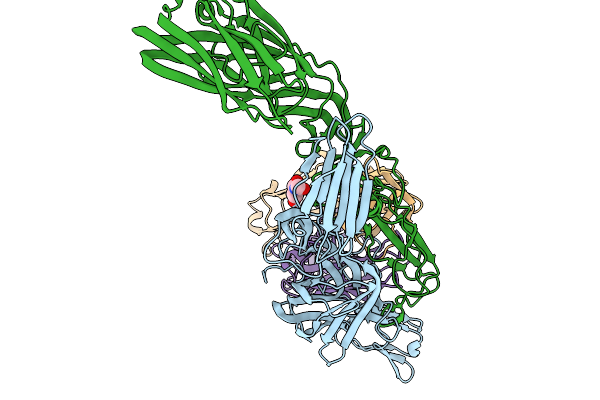
Organism: Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: VIRUS |
 |
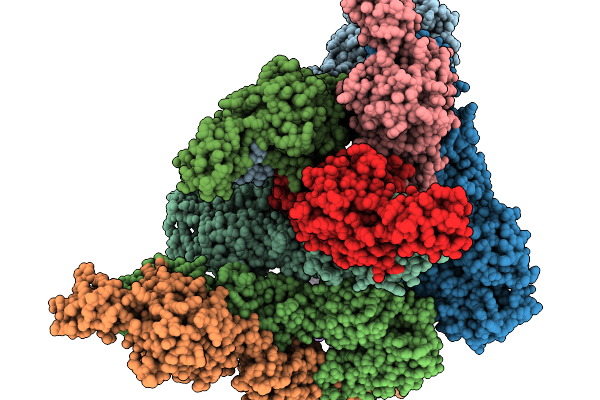
Organism: Chikungunya virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTIFUNGAL PROTEIN Ligands: NAG |
 |
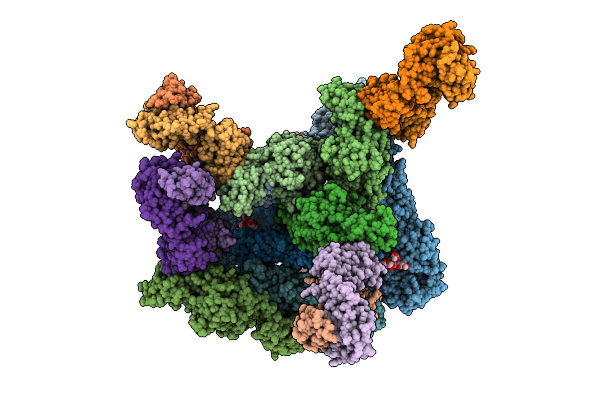
Organism: Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens, Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRUS LIKE PARTICLE Ligands: CLR |
 |
 |
Organism: Chikungunya virus, Homo sapiens
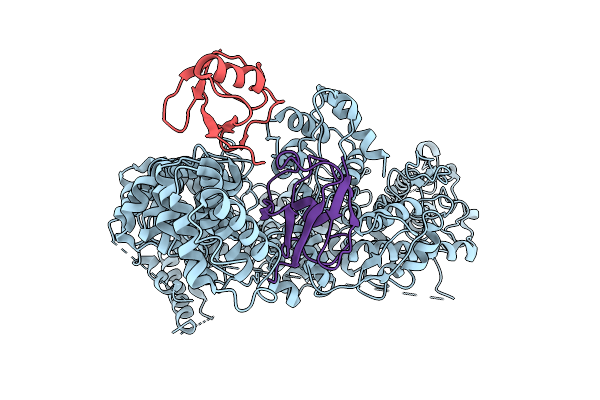
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ANTIVIRAL PROTEIN |
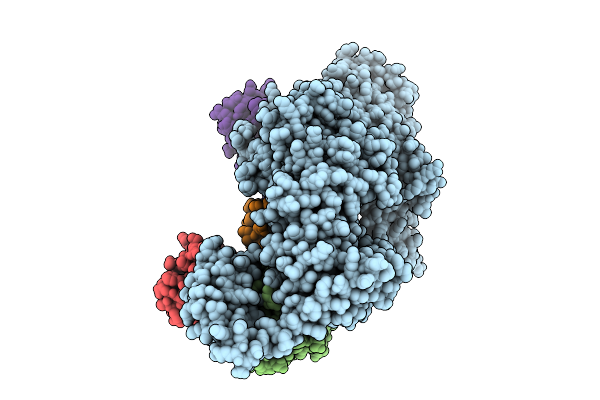 |
Cryo-Em Structure Of Ufd2/Ubc4-Ub In Complex With K29-Linked Diub (Monomeric Conformation)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
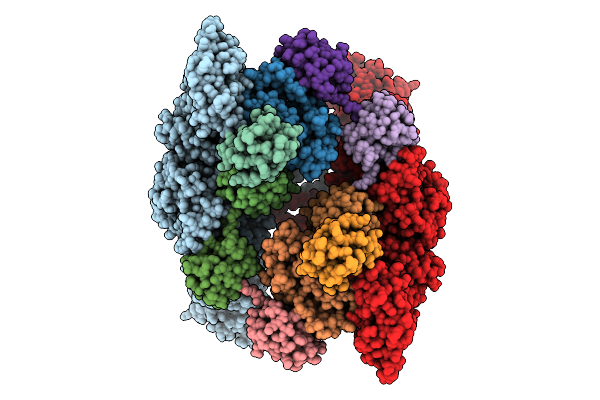 |
Cryo-Em Structure Of Ufd2/Ubc4-Ub In Complex With K29-Linked Triub (Dimeric Conformation)
Organism: Homo sapiens, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
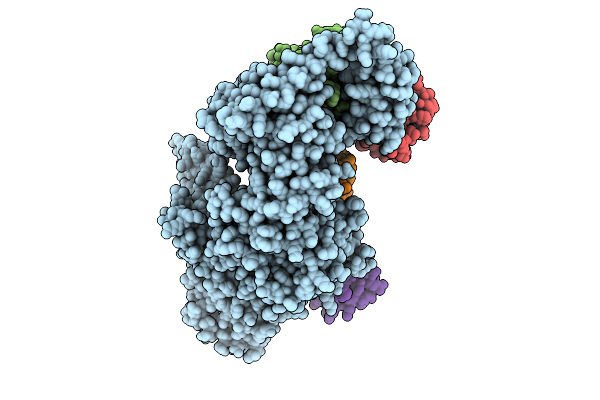 |
Cryo-Em Structure Of Ufd2/Ubc4-Ub Complex With K29Triub(Monomeric Conformation)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
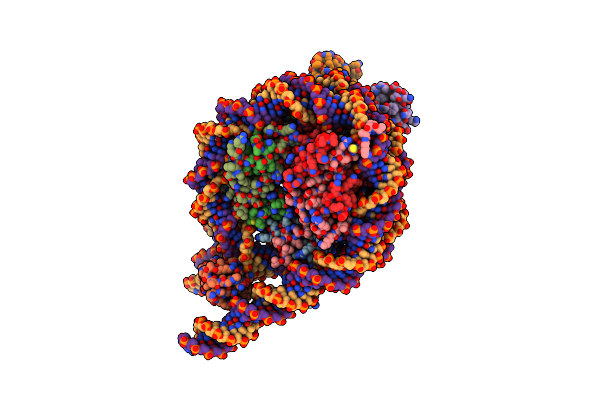 |
Cryo-Em Structure Of The Rnf168(1-193)/Ubch5C-Ub Ubiquitylation Module Bound To H1.0-K63-Ub3 Modified Chromatosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: NUCLEAR PROTEIN Ligands: ZN |
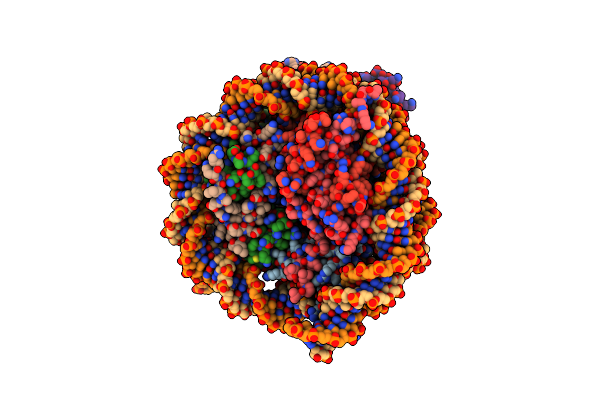 |
Cryo-Em Structures Of Rnf168/Ubch5C-Ub In Complex With H2Ak13Ub Nucleosomes Determined By Intein-Based E2-Ub-Ncp Conjugation Strategy
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: NUCLEAR PROTEIN/DNA Ligands: ZN |
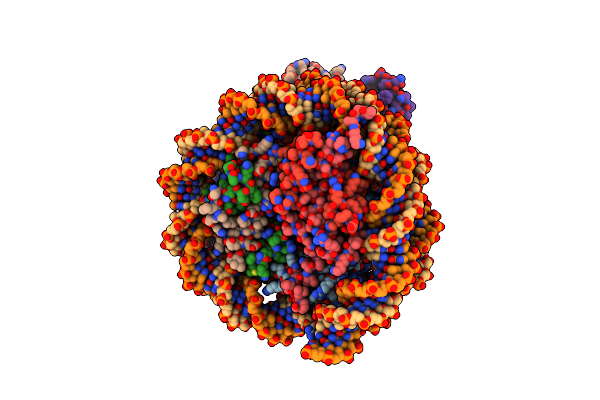 |
Cryo-Em Structures Of Rnf168/Ubch5C-Ub/Nucleosomes Complex Determined By Activity-Based Chemical Trapping Strategy
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: NUCLEAR PROTEIN/DNA Ligands: ZN |
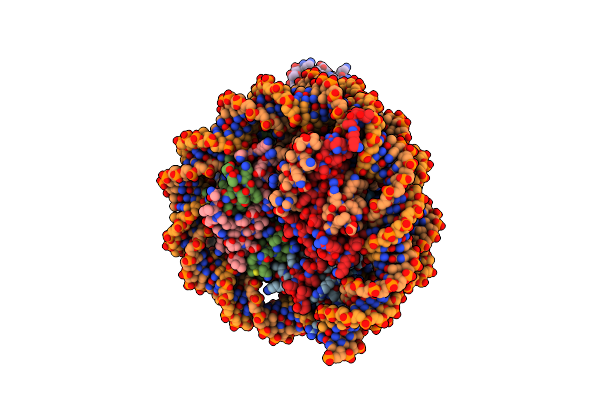 |
Cryo-Em Structures Of Rnf168/Ubch5C-Ub In Complex With H2Ak13Ub Nucleosomes Determined By Activity-Based Chemical Trapping Strategy (Adjacent H2Ak13/15 Dual-Monoubiquitination)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: NUCLEAR PROTEIN/DNA Ligands: ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/HYDROLASE/IMMUNE SYSTEM Ligands: NAG, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/HYDROLASE/IMMUNE SYSTEM Ligands: NAG, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: IMMUNE SYSTEM Ligands: NAG |

