Search Count: 35
 |
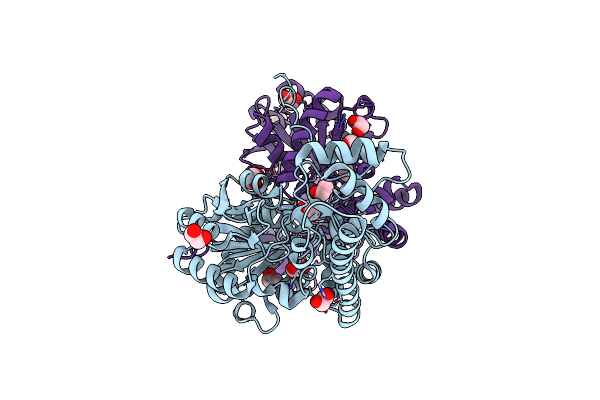
Crystal Structure Of A Glutathione S-Transferase Class Gtt2 Of Vibrio Parahaemolyticus (Vpgstt2)
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-07-07 Classification: TRANSFERASE Ligands: PEG, SO4 |
 |
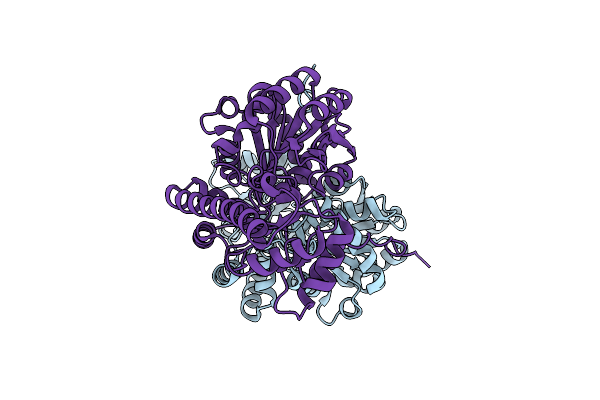
A Theoretical Optimized Mutant For The Conversion Of Substrate Specificity And Activity Of Aspartate Aminotransferase To Tyrosine Aminotransferase: Chimera P7.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-05-15 Classification: TRANSFERASE Ligands: EDO |
 |
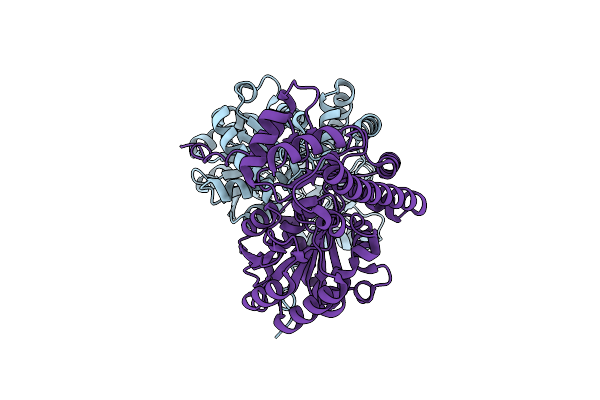
Structure Of Aspartate Aminotransferase Conversion To Tyrosine Aminotransferase: Chimera P1.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-02-13 Classification: TRANSFERASE |
 |
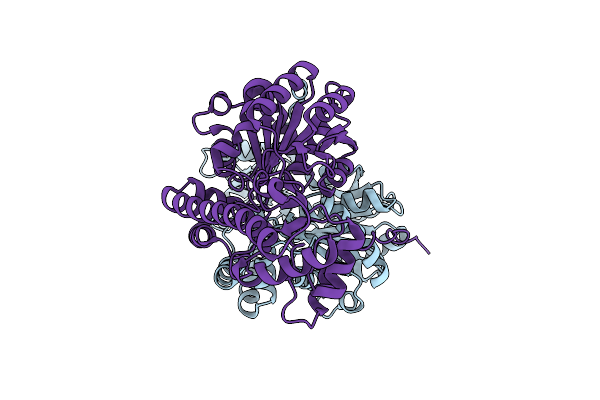
Rational Design And Directed Evolution Of E. Coli Apartate Aminotransferase To Tyrosine Aminotransferase: Mutant P2.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-02-13 Classification: TRANSFERASE |
 |
Intercoversion Of Substrate Specificity: E. Coli Aspatate Aminotransferase To Tyrosine Aminotransferase: Chimera P3.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-02-13 Classification: TRANSFERASE |
 |
Substrate Specificity Conversion Of E. Coli Pyridoxal-5'-Phosphate Dependent Aspartate Aminotransferase To Tyrosine Aminotransferase: Chimera P4.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-02-13 Classification: TRANSFERASE Ligands: MPD |
 |
Rational Design And Directed Evolution For Conversion Of Substrate Specificity From E.Coli Aspartate Aminotransferase To Tyrosine Aminotransferase: Mutant P5.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-13 Classification: TRANSFERASE |
 |
Substrate Specificity Conversion Of Aspartate Aminotransferase To Tyrosine Aminotransferase By The Janus Algorithm: Chimera P6.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-02-13 Classification: TRANSFERASE |
 |
Wild-Type E. Coli Aspartate Aminotransferase: A Template For The Interconversion Of Substrate Specificity And Activity To Tyrosine Aminotransferase By The Janus Algorithm.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-02-13 Classification: TRANSFERASE Ligands: EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-12-05 Classification: TRANSFERASE Ligands: SO4, EDO, 3QP |
 |
Crystal Structures Of Escherichia Coli Aspartate Aminotransferase Reconstituted With 1-Deaza-Pyridoxal 5'-Phosphate: Internal Aldimine And Stable L-Aspartate External Aldimine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-06-15 Classification: TRANSFERASE Ligands: SO4, EDO |
 |
Crystal Structures Of Escherichia Coli Aspartate Aminotransferase Reconstituted With 1-Deaza-Pyridoxal 5'-Phosphate: Internal Aldimine And Stable L-Aspartate External Aldimine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-06-15 Classification: TRANSFERASE Ligands: SO4, EDO, CL, 3QP |
 |
Crystal Structure Of Putative Aspartate Aminotransferase (Np_905498.1) From Porphyromonas Gingivalis W83 At 1.75 A Resolution
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-02-10 Classification: TRANSFERASE |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2006-08-08 Classification: LYASE |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-08-08 Classification: LYASE Ligands: CS, NA, MES, PLP |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-01-03 Classification: LYASE |
 |
The Crystal Structure Of Dialkylglycine Decarboxylase Complex With Pyridoxamine 5-Phosphate
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-01-03 Classification: LYASE Ligands: K, NA, PMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2005-03-01 Classification: TRANSFERASE Ligands: SO4, EDO, PMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-03-01 Classification: TRANSFERASE Ligands: SO4, EDO, PLP, PMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2005-03-01 Classification: TRANSFERASE Ligands: SO4, EDO, PLP, PMP |

