Search Count: 29
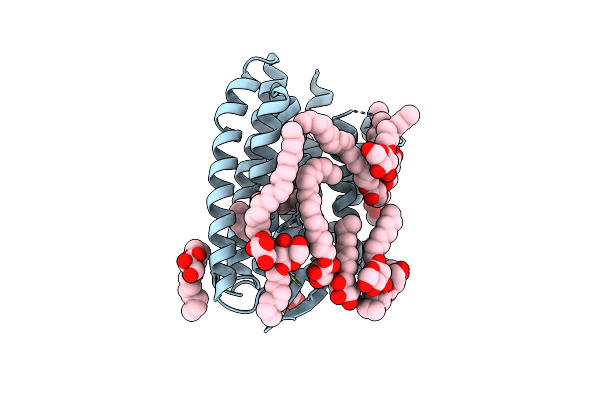 |
Grouped 150-240 Ms Dark Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
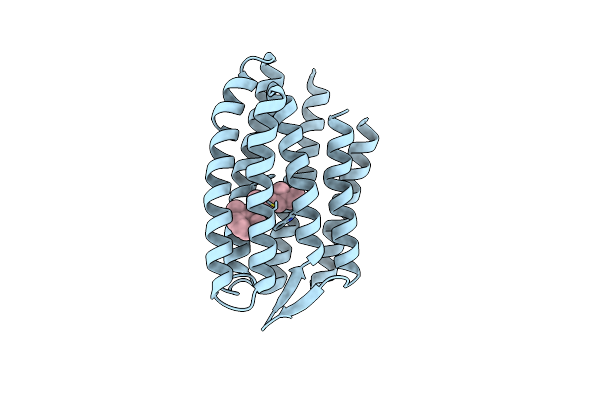 |
Continuously Illuminated Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL |
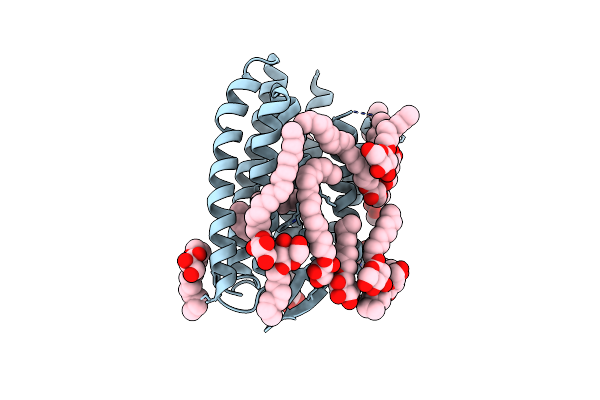 |
Continuous Dark State Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
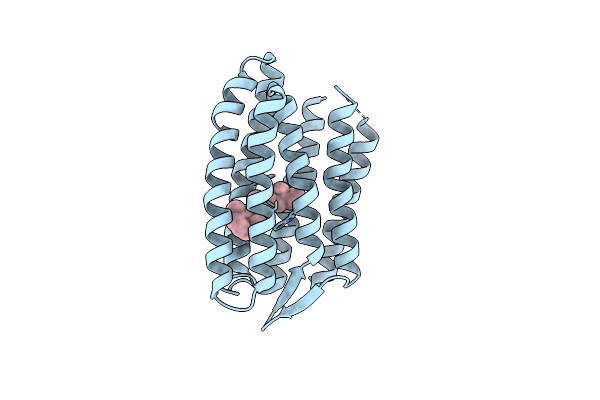 |
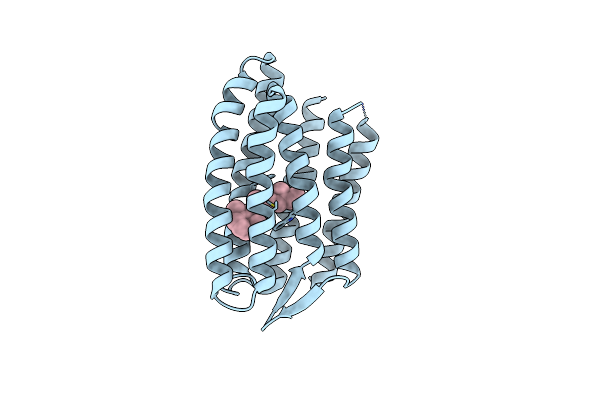
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 90-120 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
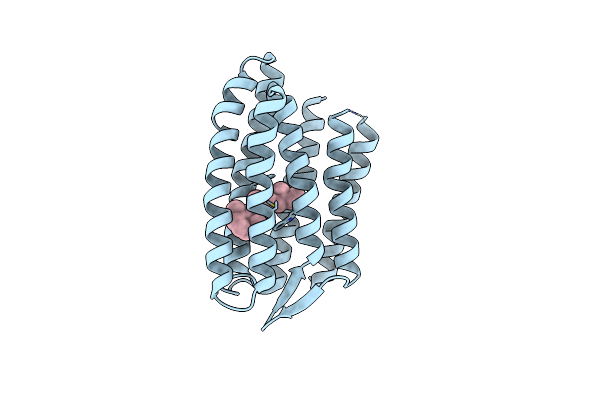
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 60-90 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography. 30-60 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 0-30 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 120-150 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
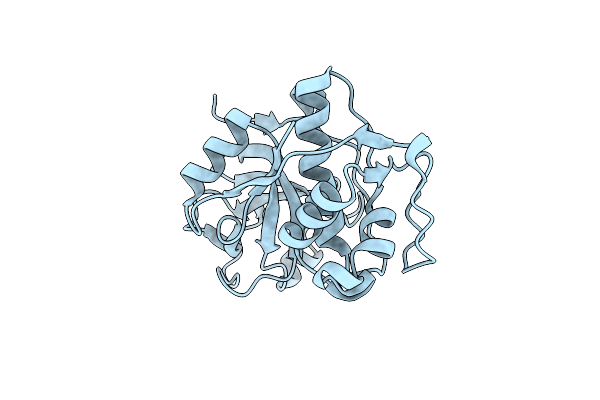 |
Organism: Tethya aurantium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-12-02 Classification: HYDROLASE |
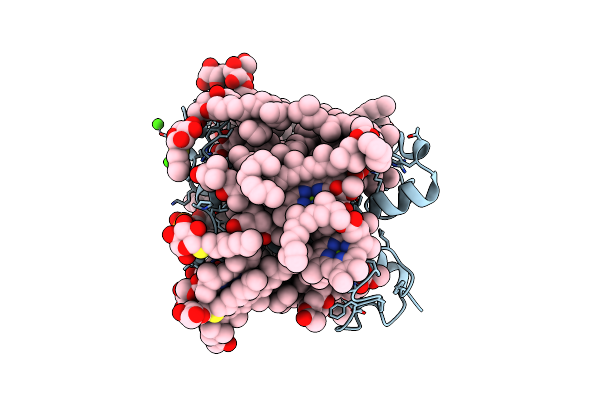 |
Crystal Structure Of Fucoxanthin Chlorophyll A/C Complex From Phaeodactylum Tricornutum
Organism: Phaeodactylum tricornutum ccap 1055/1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-02-06 Classification: PHOTOSYNTHESIS Ligands: CA, A86, DD6, CLA, KC2, KC1, LMT, DGD, LHG, SOG, UNL |
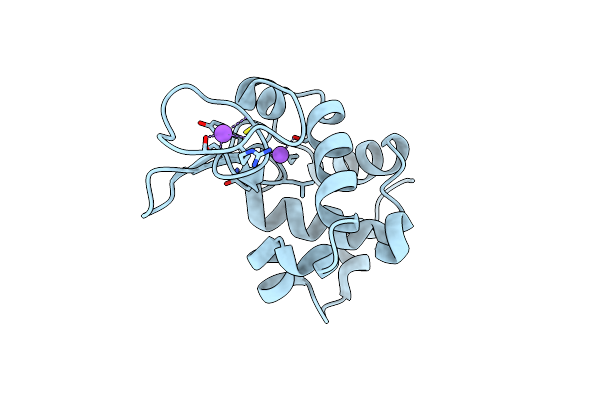 |
Hen Egg Lysozyme At Room Temperature Solved From Datasets Acquired By Ultrasonic Acoustic Levitation Method And Processed By Crystfel
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: CL, NA |
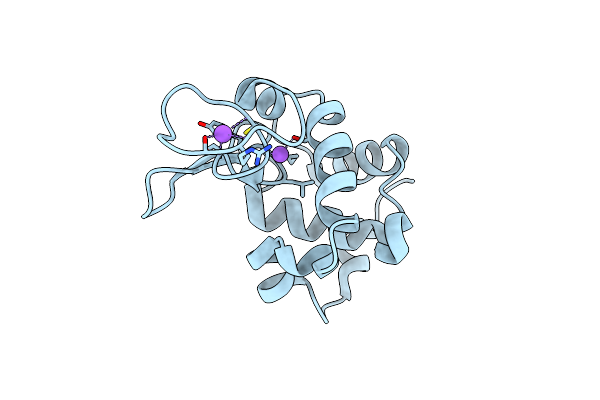 |
Hen Egg Lysozyme At Room Temperature Solved From 3600 Diffraction Images Acquired By Ultrasonic Acoustic Levitation Method And Processed By Crystfel
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: CL, NA |
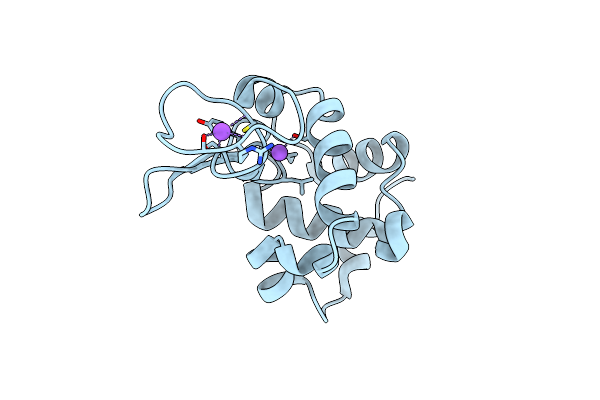 |
Hen Egg Lysozyme At Room Temperature Solved From Dataset Acquired By Oscillation Method
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: CL, NA |
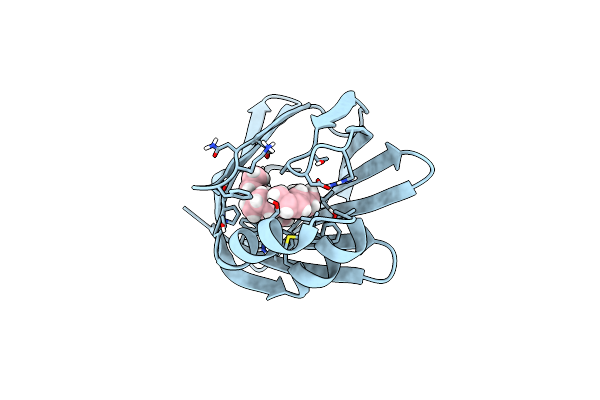 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:0.98 Å Release Date: 2016-03-09 Classification: CELL CYCLE Ligands: OLA |
 |
Organism: Mus musculus, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-08-06 Classification: TRANSPORT PROTEIN Ligands: CL, SO4, NAG |
 |
The 1.3 A Resolution Structure Of Nitrosomonas Europaea Rh50 And Mechanistic Implications For Nh3 Transport By Rhesus Family Proteins
Organism: Nitrosomonas europaea
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2007-11-20 Classification: TRANSPORT PROTEIN Ligands: BOG, GOL |
 |
Crystal Structure Of Aldose Reductase With Citrates Bound In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2004-09-07 Classification: OXIDOREDUCTASE Ligands: NAP, CIT |
 |
Crystal Structure Of Aldose Reductase Complexed With 2R4S (Stereoisomer Of Fidarestat, 2S4S)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2004-09-07 Classification: OXIDOREDUCTASE Ligands: NAP, FIR |
 |
Crystal Structure Of Aldose Reductase Complexed With 2S4R (Stereoisomer Of Fidarestat, 2S4S)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2004-09-07 Classification: OXIDOREDUCTASE Ligands: NAP, CIT, FIS |
 |
Crystal Structure Of Human Aldose Reductase Complexed With Nadp And Minalrestat
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2004-02-24 Classification: OXIDOREDUCTASE Ligands: BFI, NAP |

