Search Count: 12
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-11-02 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: NAG, 6VR, PEG |
 |
Structure-Based Design Of A New Series Of N-Piperidin-3-Ylpyrimidine-5-Carboxamides As Renin Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-11-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 74U, PEG, NAG, PGE |
 |
Structure-Based Design Of A New Series Of N-Piperidin-3-Ylpyrimidine-5-Carboxamides As Renin Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-11-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG, 74V, PGE, DMS, 1PE |
 |
Structure-Based Design Of A New Series Of N-Piperidin-3-Ylpyrimidine-5-Carboxamides As Renin Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-11-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG, 74Z, DMS, PEG, 1PE |
 |
Structure-Based Design Of A New Series Of N-Piperidin-3-Ylpyrimidine-5-Carboxamides As Renin Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2016-11-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG, 74Y, PEG, PGE |
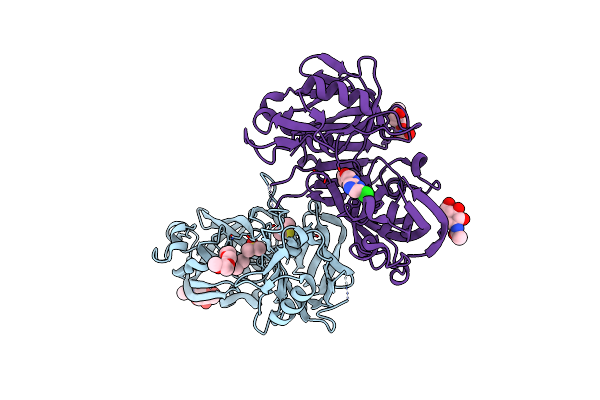 |
Novel Approach Of Fragment-Based Lead Discovery Applied To Renin Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2016-10-26 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: NAG, PG6, DMS, 77O, PGE |
 |
Crystal Structure Of Hepatitis C Virus Rna-Dependent Rna Polymerase Ns5B In Complex With Small Molecule Fragments
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: ZN, SX1 |
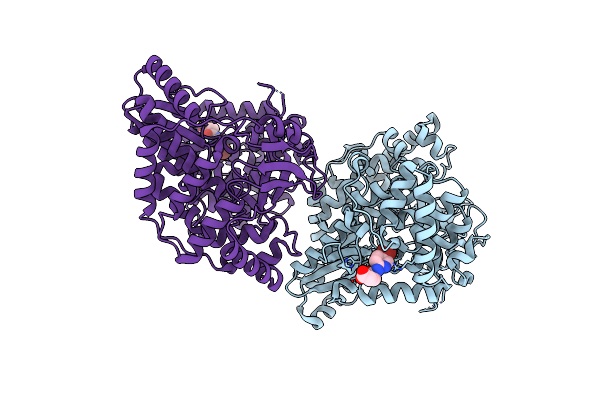 |
Crystal Structure Of Hepatitis C Virus Rna-Dependent Rna Polymerase Ns5B In Complex With Small Molecule Fragments
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: SX2 |
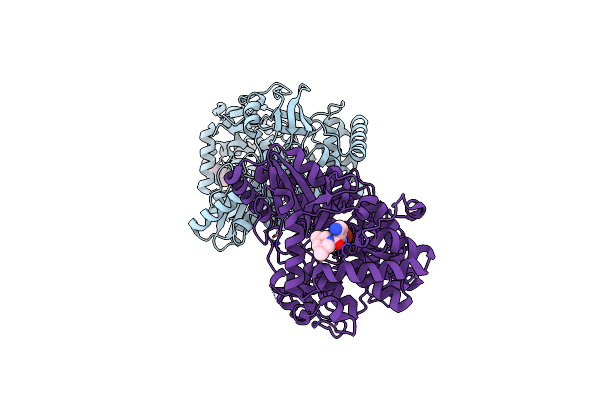 |
Crystal Structure Of Hepatitis C Virus Rna-Dependent Rna Polymerase Ns5B In Complex With Optimized Small Molecule Fragments
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: NI, SX3 |
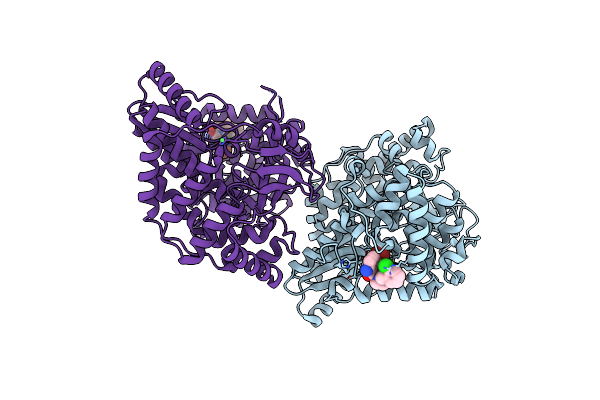 |
Crystal Structure Of Hepatitis C Virus Rna-Dependent Rna Polymerase Ns5B In Complex With Optimized Small Molecule Fragments
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: NI, SX4 |
 |
Crystal Structure Of Hepatitis C Virus Rna-Dependent Rna Polymerase Ns5B In Complex With Optimized Small Molecule Fragments
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: NI, SX5 |
 |
Crystal Structure Of Hepatitis C Virus Rna-Dependent Rna Polymerase Ns5B In Complex With Optimized Small Molecule Fragments
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: SX6 |

