Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
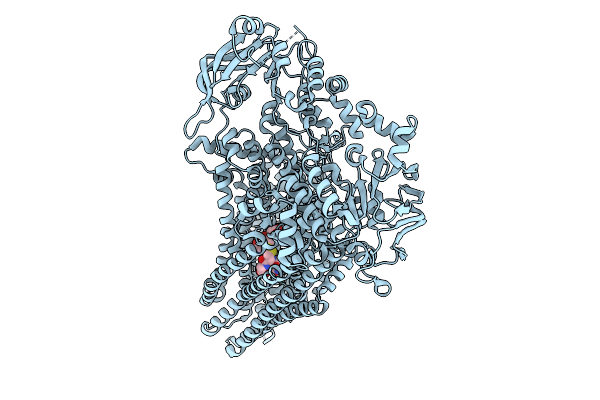 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: GDS |
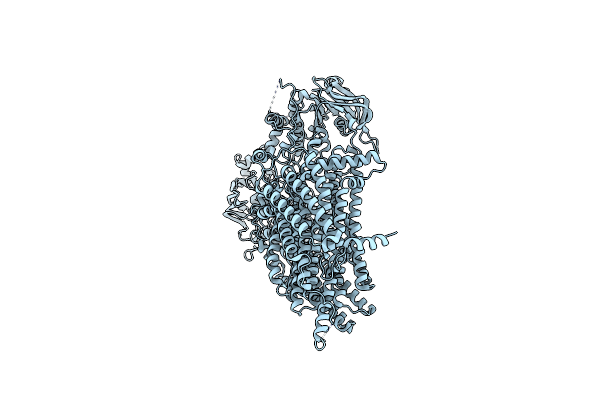 |
Tetra-Phosphorylated, E1435Q Ycf1 Mutant In Inward-Facing Wide Conformation
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: MEMBRANE PROTEIN |
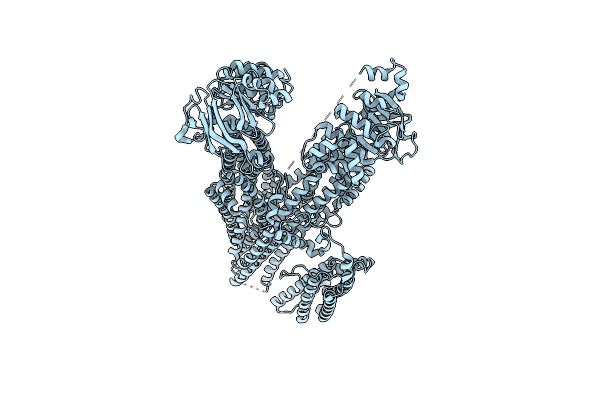 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: MEMBRANE PROTEIN |
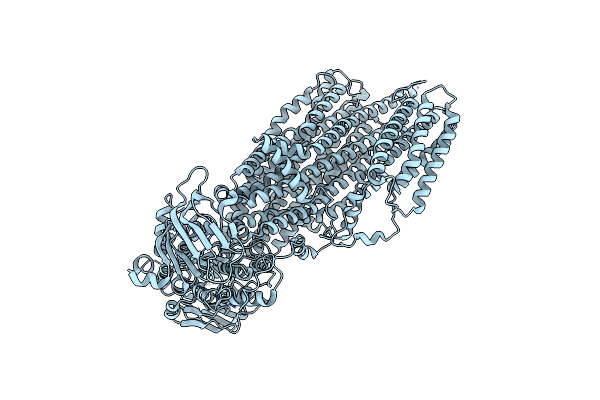 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
 |
The Structure Of Bacillus Subtilis Bmrcd In The Inward-Facing Conformation Bound To Hoechst-33342 And Atp
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2022-01-05 Classification: TRANSPORT PROTEIN Ligands: ATP, HT1 |
 |
Escherichia Coli Quinol:Fumarate Reductase Crystallized Without Dicarboxylate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2017-12-06 Classification: ELECTRON TRANSPORT Ligands: FAD, PO4, FES, F3S, SF4 |
 |
Crystal Structure Of The Heterodimeric Abc Transporter Tmrab, A Homolog Of The Antigen Translocation Complex Tap
Organism: Thermus thermophilus (strain hb27 / atcc baa-163 / dsm 7039)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-01-18 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Organism: Streptococcus gordonii
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2016-04-13 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of Lanosterol 14-Alpha Demethylase With Intact Transmembrane Domain Bound To Itraconazole
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2016-01-13 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: HEM, 1YN |
 |
Saccharomyces Cerevisiae Lanosterol 14-Alpha Demethylase With Lanosterol Bound
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, LAN, OXY |
 |
Plasticity Of The Quinone-Binding Site Of The Complex Ii Homolog Quinol:Fumarate Reductase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-07-17 Classification: OXIDOREDUCTASE Ligands: FAD, FES, F3S, SF4, MQ7 |
 |
Organism: Streptococcus gordonii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-08-10 Classification: SUGAR BINDING PROTEIN Ligands: K, GOL, HDZ, NA |
 |
Organism: Streptococcus gordonii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-08-10 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL, HDZ |
 |
Crystal Structure Of Menaquinol:Oxidoreductase In Complex With Oxaloacetate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, OAA, FES, F3S, SF4 |
 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With A 3-Nitropropionate Adduct
Organism: Escherichia coli 042, Escherichia coli o55:h7 str. cb9615
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE Ligands: FAD, 3NP, FES, F3S, SF4 |
 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Fumarate
Organism: Escherichia coli 042, Escherichia coli 536, Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-12-01 Classification: OXIDOREDUCTASE Ligands: FAD, FUM, FES, F3S, SF4 |
 |
Crystal Structure Of Menaquinol:Fumarate Oxidoreductase In Complex With Glutarate
Organism: Escherichia coli 042, Escherichia coli o55:h7
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2010-11-24 Classification: OXIDOREDUCTASE Ligands: FAD, GUA, FES, F3S, SF4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2008-04-01 Classification: OXIDOREDUCTASE Ligands: FAD, FES, F3S, SF4 |