Search Count: 12
 |
Glucagon Like Peptide Receptor-1 (Glp1R) A316T Mutant With Glp-1 Peptide. Dominant Negative Gs Complex.
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN |
 |
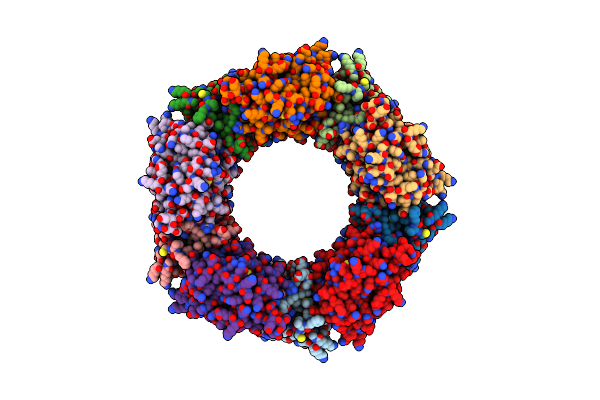
Mitochondrial Peroxiredoxin From Leishmania Infantum In Complex With Unfolding Client Protein After Heat Stress
Organism: Leishmania infantum
Method: ELECTRON MICROSCOPY Release Date: 2019-02-20 Classification: CHAPERONE |
 |
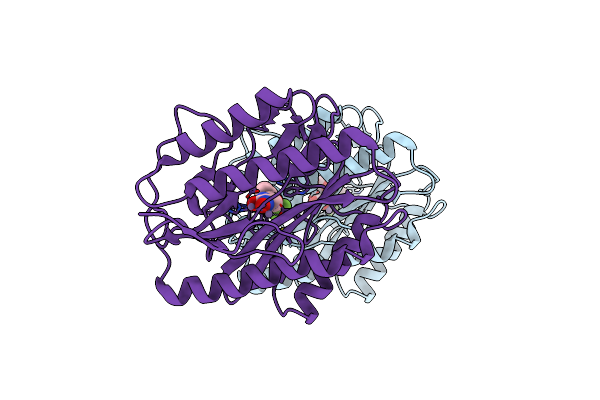
Mitochondrial Peroxiredoxin From Leishmania Infantum After Heat Stress Without Unfolding Client Protein
Organism: Leishmania infantum
Method: ELECTRON MICROSCOPY Release Date: 2019-02-20 Classification: CHAPERONE |
 |
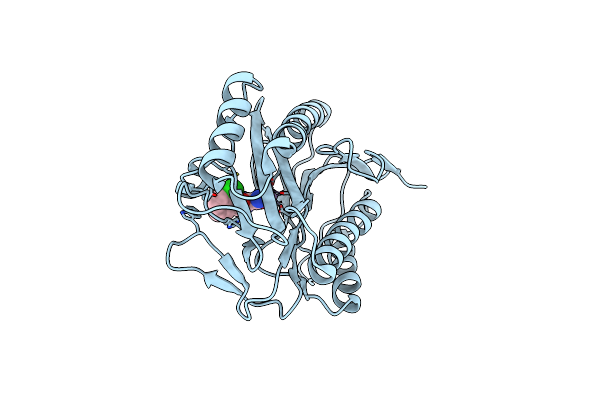
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2012-06-13 Classification: HYDROLASE Ligands: MN, CO3, IKY |
 |
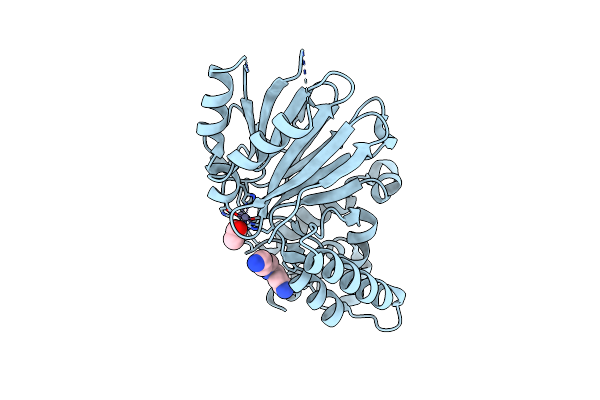
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2012-06-13 Classification: TRANSFERASE Ligands: MN, 5C1 |
 |
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-01-15 Classification: HYDROLASE Ligands: ZN, SPD, ACY |
 |
Crystal Structure Of The Leishmania Infantum Glyoxalase Ii With D-Lactate At The Active Site
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-01-15 Classification: HYDROLASE Ligands: ZN, SPD, LAC |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-03-25 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, CA, 0PQ, DMS |
 |
The 2.2 A Resolution Structure Of Thermolysin Crystallized In Presence Of Potassium Thiocyanate
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2002-12-05 Classification: HYDROLASE Ligands: VAL, LYS, CA, ZN, SCN |
 |
Thermolysin (E.C.3.4.24.27) Complexed With (2-Sulphanyl-3-Phenylpropanoyl)-Phe-Tyr. Parameters For Zn-Bidentation Of Mercaptoacyldipeptides In Metalloendopeptidase
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-12-29 Classification: HYDROLASE Ligands: ZN, CA, TI2, DMS |
 |
Thermolysin (E.C.3.4.24.27) Complexed With (2-Sulphanylheptanoyl)-Phe-Ala. Parameters For Zn-Bidentation Of Mercaptoacyldipeptides In Metalloendopeptidase
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-12-29 Classification: HYDROLASE Ligands: ZN, CA, TI1, DMS |
 |
Thermolysin (E.C.3.4.24.27) Complexed With (2-Sulphanyl-3-Phenylpropanoyl)-Gly-(5-Phenylproline). Parameters For Zn-Monodentation Of Mercaptoacyldipeptides In Metalloendopeptidase
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 1999-12-29 Classification: HYDROLASE Ligands: ZN, CA, TI3, DMS |

