Search Count: 72
 |
Sars-Cov-2 Papain-Like-Protease (Plpro) In Complex With Inhibitor Linagliptin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: 356, GOL |
 |
Dengue 3 Ns5 Methyltransferase Bound To S-Adenosyl-L-Homocysteine And Herbacetin
Organism: Dengue virus type 3
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: SAH, A1L1Z, GOL |
 |
Sars-Cov-2 Papain Like Protease (Plpro) In Complex With Inhibitor Lithocholic Acid
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: 4OA, GOL, NA |
 |
Crystal Structure Of C-Terminal Domain Of Nucleocapsid Protein From Sars-Cov-2 In Complex With Ceftriaxone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-02 Classification: VIRAL PROTEIN Ligands: SO4, 9F2, PEG |
 |
Crystal Structure Of C-Terminal Domain Of Nucleocapsid Protein From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-02 Classification: VIRAL PROTEIN |
 |
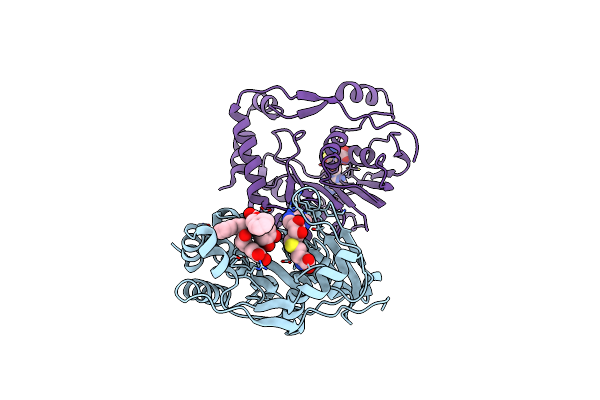
Crystal Structure Of C-Terminal Domain Of Nucleocapsid Protein From Sars-Cov-2 In Complex With Ampicillin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: EDO, AIC |
 |
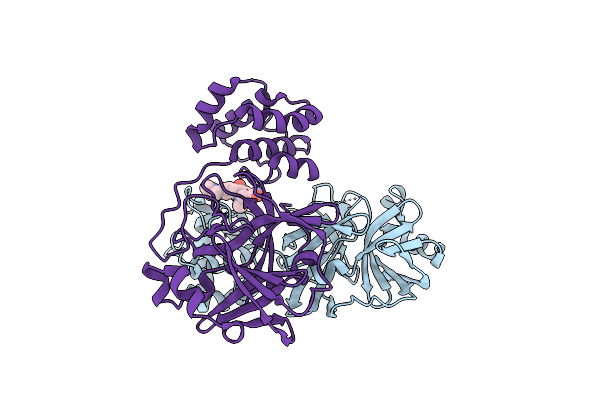
Dengue 3 Ns5 Methyltransferase Bound To S-Adenosyl-L-Homocysteine And Caffeic Acid Phenethyl Ester
Organism: Dengue virus type 3
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN Ligands: SAH, QAP |
 |
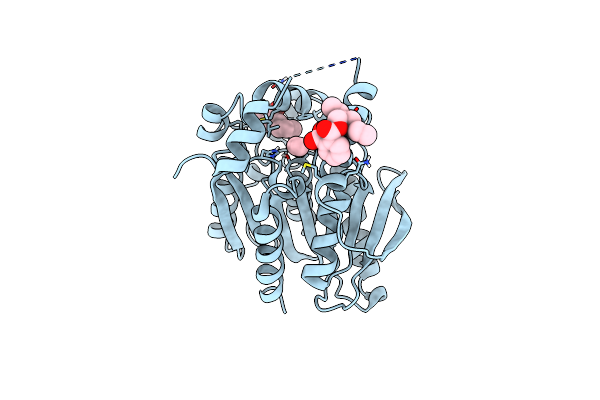
The Crystal Structure Of Covid-19 Main Protease In Complex With An Inhibitor Minocycline
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-02-12 Classification: ANTIVIRAL PROTEIN Ligands: MIY |
 |
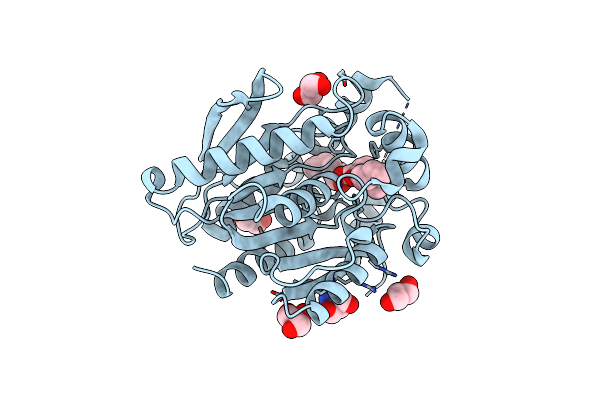
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Monoethylhexyl Phtahalate And 2-Ethylhexanol
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: QGL, ACE, 2EH |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus S154A Mutant In Complex With Diethylhexyl Phthalate At 2.4A
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: TKU, TRS, GOL |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus At 1.22A
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, MLI, ACE |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Phthalate
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, PHT |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Monomethyl Phthalate
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: R3O, EDO |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Paranitrophenyl Butyrate
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, R5C |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Jeffamine
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, JEF, MLA |
 |
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With P-Nitrophenol
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, NPO, ACE |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Dimethyl Phthalate
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: TIK, EDO, ACE |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Diethylhexyl Phthalate
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: TKU |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Dimethyl Phthalate On Surface
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: TIK, EDO, MLA, ACE |

