Search Count: 14
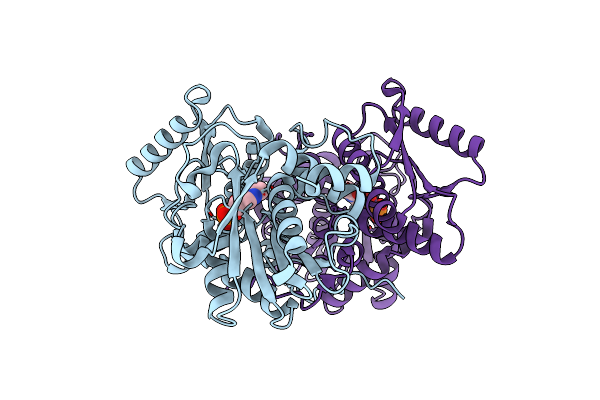 |
Crystal Structure Of O-Acetyl Serine Sulfhydrylase Isoform 3 From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: PLP |
 |
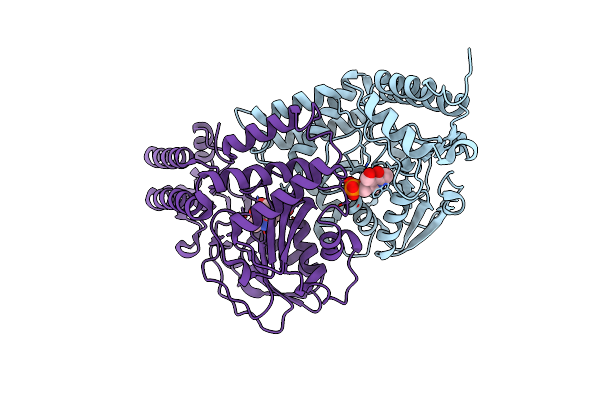
Crystal Structure Of Wild Type Phosphoserine Aminotransferase (Psat) From E. Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Release Date: 2018-10-31 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Crystal Structure Of Delta 4 Mutant Of Ehpsat (Phosphoserine Aminotransferase Of Entamoeba Histolytica)
Organism: Entamoeba histolytica hm-3:imss
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-10-31 Classification: TRANSFERASE Ligands: CL, PLP |
 |
Crystal Structure Of 45 Amino Acid Deleted From N-Terminal Of Phosphoserine Aminotransferase (Psat) Of Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: SCN |
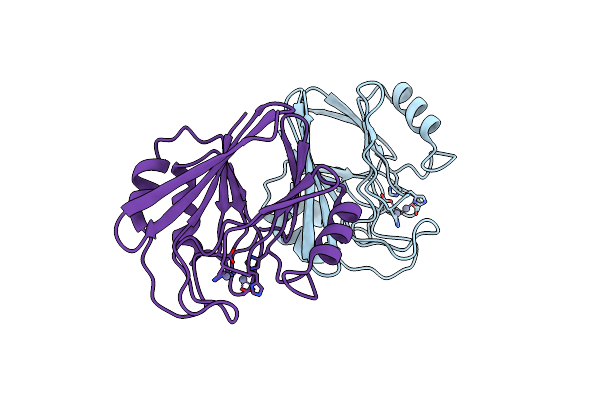 |
Crystal Structure Of Wild Type Vps29 Complexed With Zn+2 From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-10-18 Classification: PROTEIN TRANSPORT Ligands: ZN |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-10-18 Classification: PROTEIN TRANSPORT |
 |
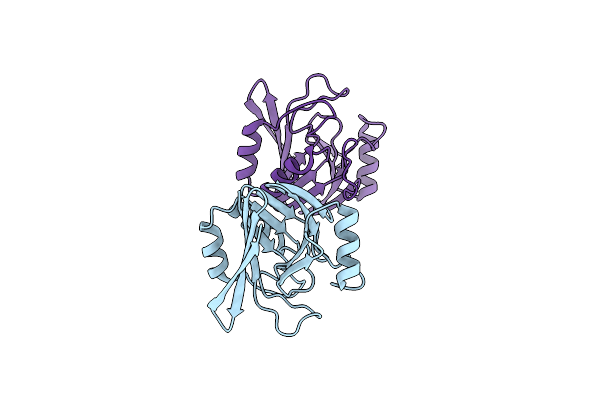
Crystal Structure Of Vps29 Double Mutant (D62A/H86A) From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-10-18 Classification: PROTEIN TRANSPORT |
 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-10-04 Classification: TRANSPORT PROTEIN |
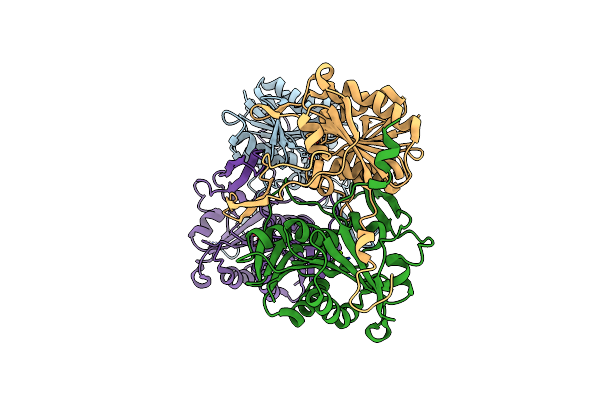 |
Structural And Functional Insights Into Survival Endonuclease, An Important Virulence Factor Of Brucella Abortus
Organism: Brucella abortus s19
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-06 Classification: HYDROLASE Ligands: MG |
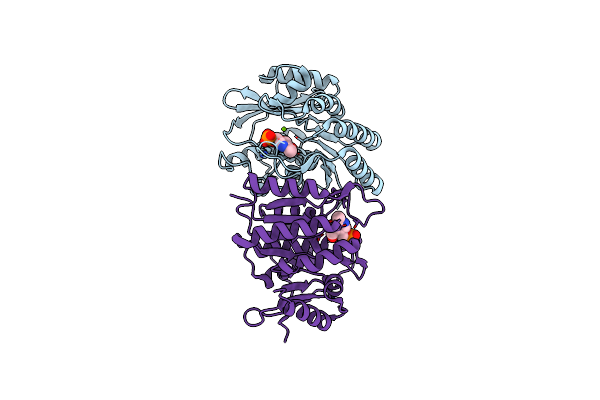 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-02-04 Classification: TRANSFERASE Ligands: PLP, MG |
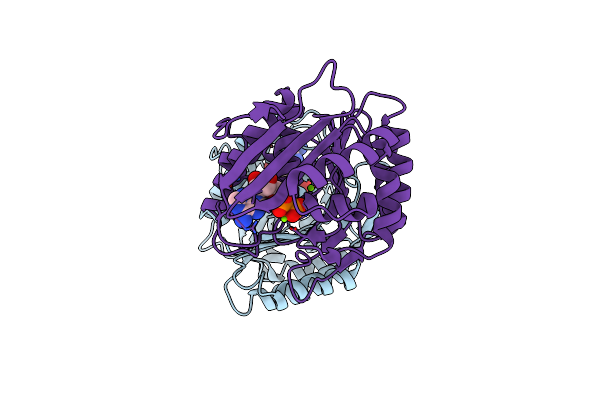 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-01-28 Classification: TRANSFERASE Ligands: ADP, MG |
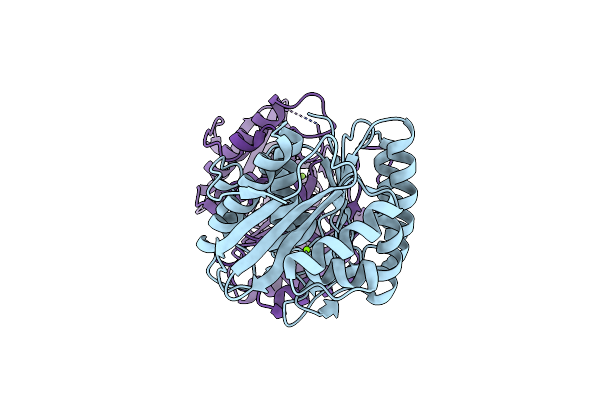 |
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2015-01-28 Classification: TRANSFERASE Ligands: MG |
 |
Crystal Structure Of Murraya Koenigii Miraculin-Like Protein At 2.2 A Resolution At Ph 7.0
Organism: Murraya koenigii
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2013-12-04 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure Of Murraya Koenigii Miraculin-Like Protein At 2.2 A Resolution At Ph 4.6
Organism: Murraya koenigii
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2013-12-04 Classification: HYDROLASE INHIBITOR |

