Search Count: 16
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE |
 |
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-03-04 Classification: HYDROLASE |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Without Soaking)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CA, CL |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-1A)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CA, CS, CL |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-1B)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CS, CA |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-1C)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: CS, CA |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-2A)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: SR, CL |
 |
N288Q-N321Q Mutant Beta-Lactamase Derived From Chromohalobacter Sp.560 (Condition-2B)
Organism: Chromohalobacter sp. 560
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: SR, CL, CA |
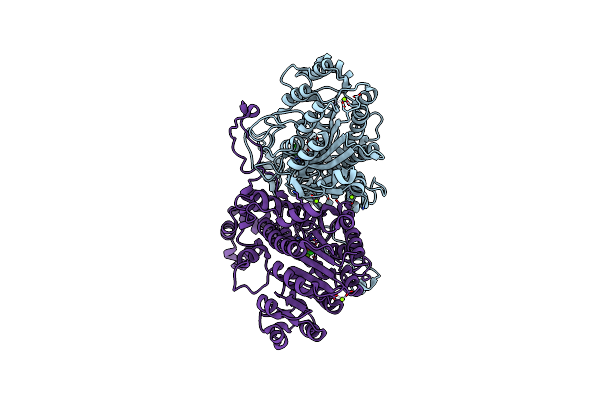 |
Structural Characteristics Of Alkaline Phosphatase From A Moderately Halophilic Bacteria Halomonas Sp.593
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-12 Classification: HYDROLASE Ligands: MG, ZN, CL |
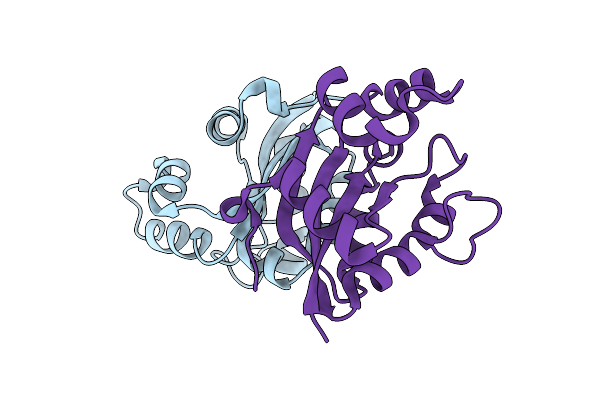 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Halomonas
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-11 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-02-07 Classification: SIGNALING PROTEIN/CYTOKINE |
 |
Structure Of A Halophilic Nucleoside Diphosphate Kinase From Halobacterium Salinarum
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2005-12-20 Classification: TRANSFERASE Ligands: CA |
 |
Structure Of A Halophilic Nucleoside Diphosphate Kinase From Halobacterium Salinarum In Complex With Cdp
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-12-20 Classification: TRANSFERASE Ligands: MG, CDP |

