Search Count: 76
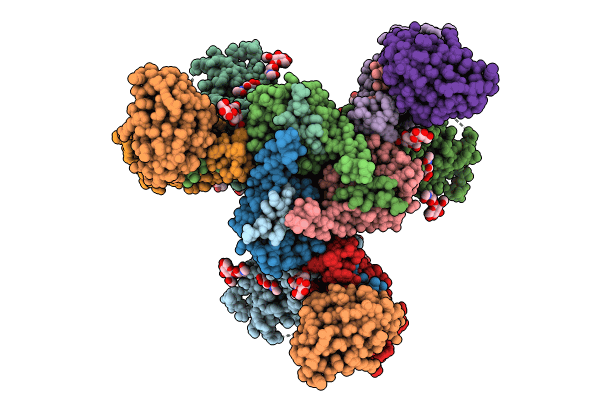 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-B.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
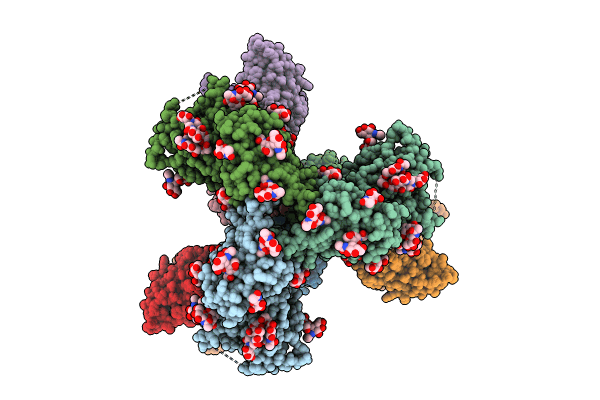
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
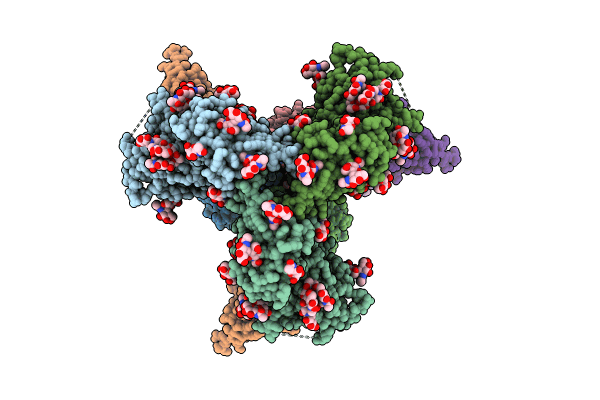
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-D.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
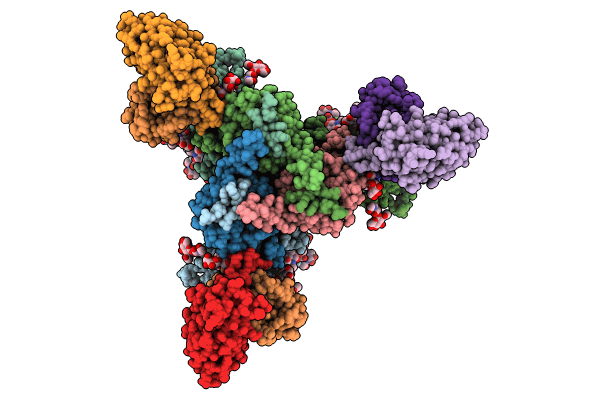
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-E.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
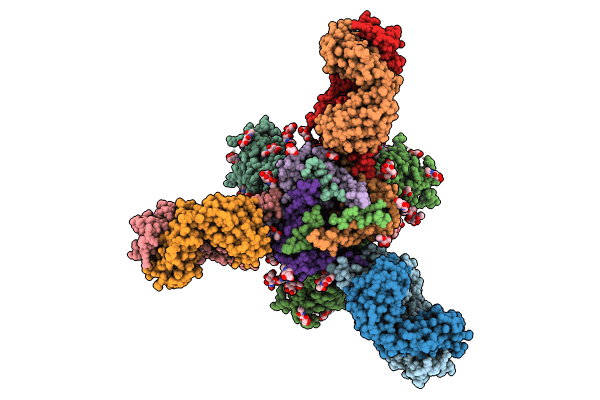
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Trnm-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Herh-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Trnm-F*01 Fab In Complex With Hiv-1 Env Trimer Conc Sosip
Organism: Homo sapiens, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
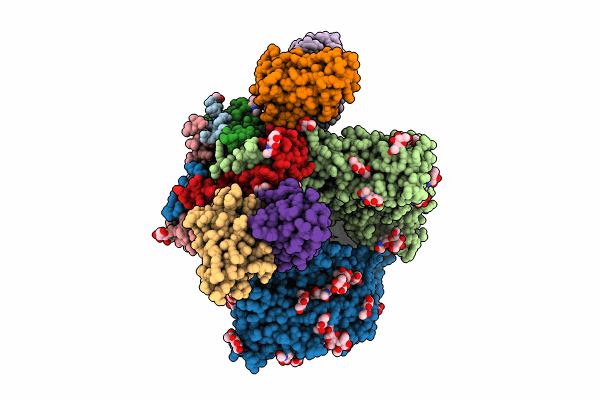
Cryo-Em Structure Of Hiv-1 Env Bg505 Ds-Sosip In Complex With Antibody Gpz6-B.01 Targeting The Fusion Peptide
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-A.01 Fab
Organism: Macaca, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: Viral Protein/Immune System Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Gpz6-A.01 Fab
Organism: Human immunodeficiency virus 1, Macaca
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: Viral PROTEIN/iMMUNE SYSTEM Ligands: NAG |
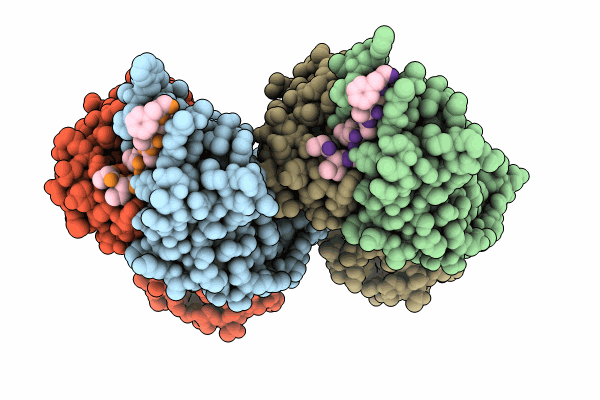 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM |
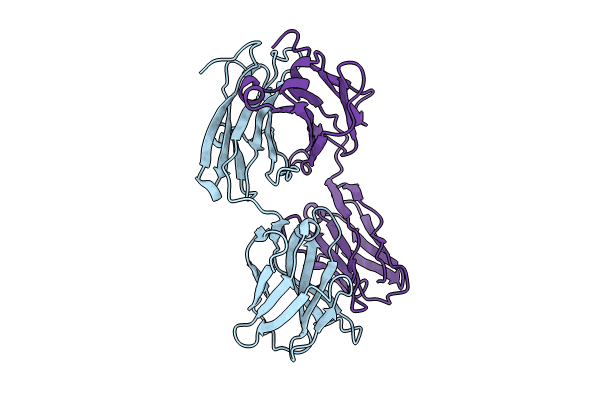 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM |
 |
Organism: Lassa virus josiah, Camelus bactrianus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
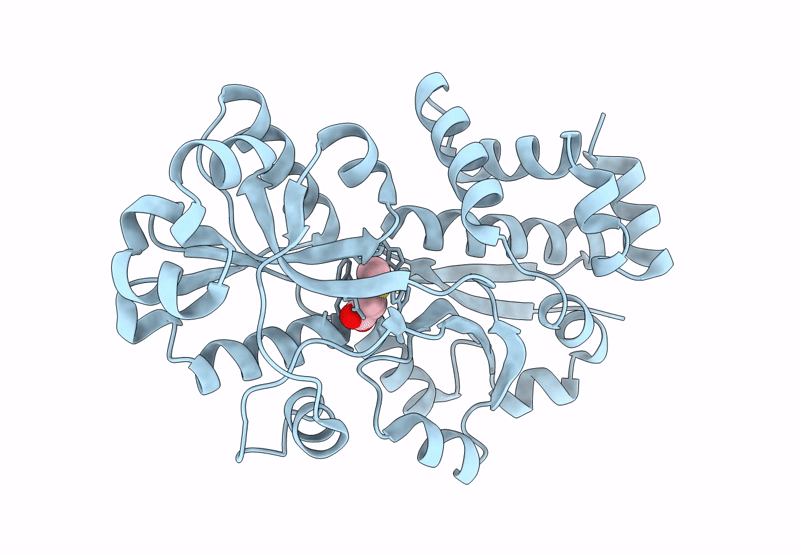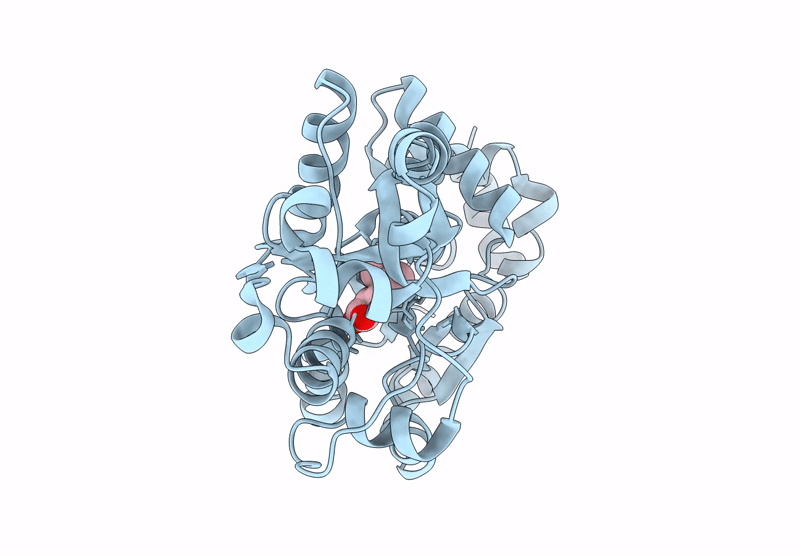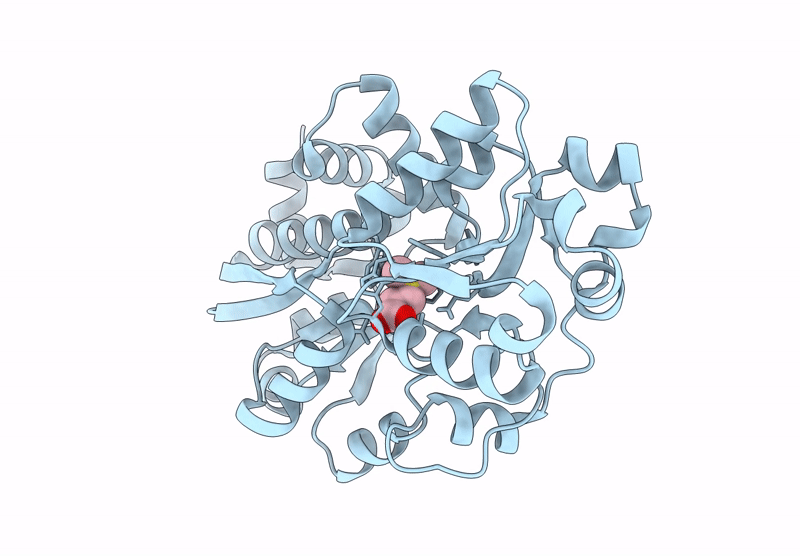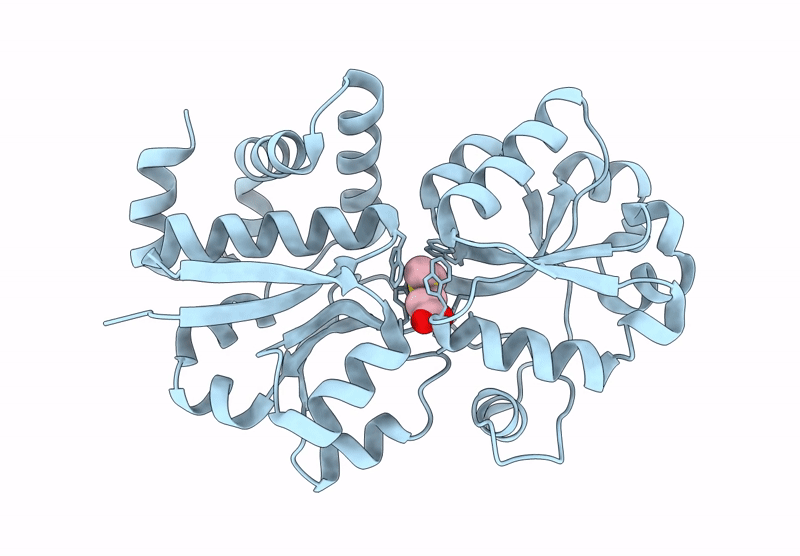
Organism: Acinetobacter bereziniae niph 3
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-10-04 Classification: LYASE Ligands: ZN, LNI |
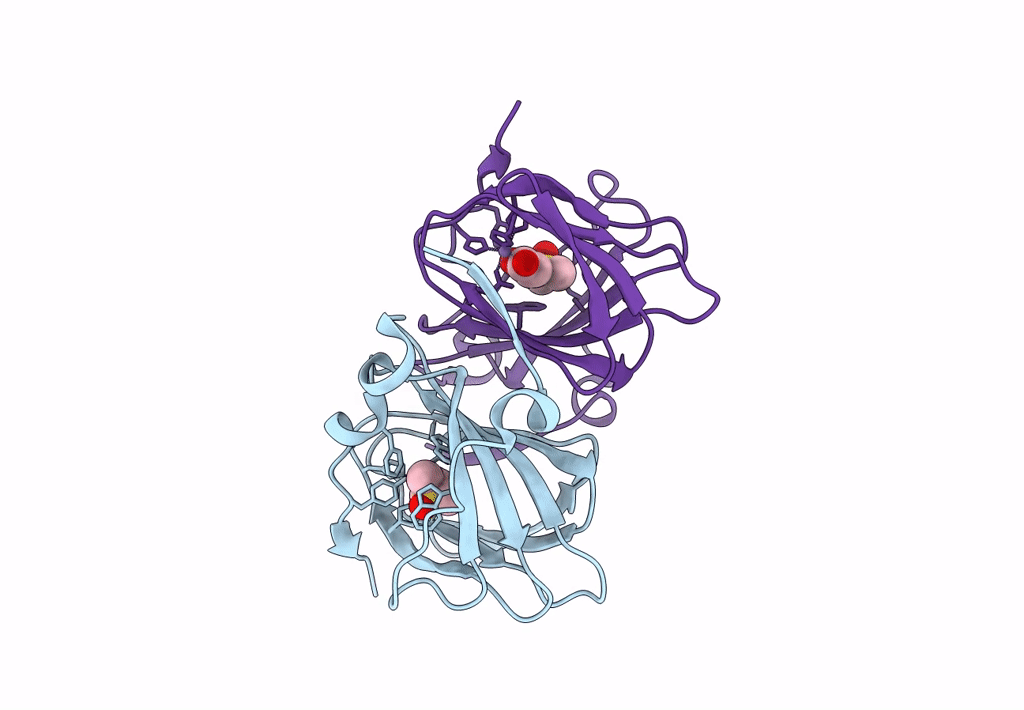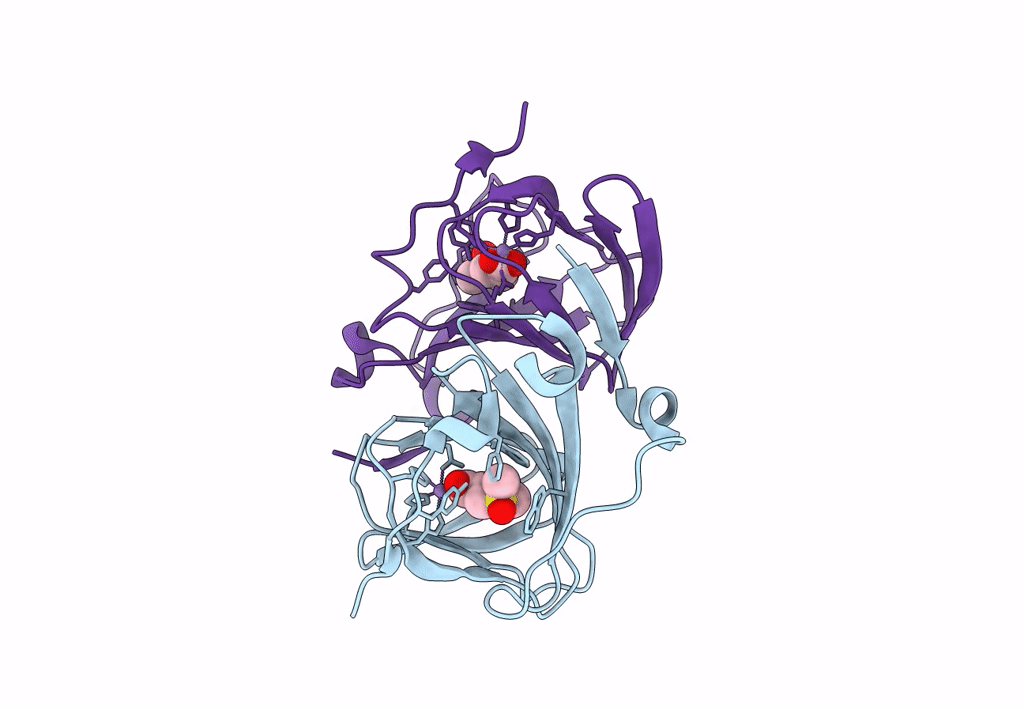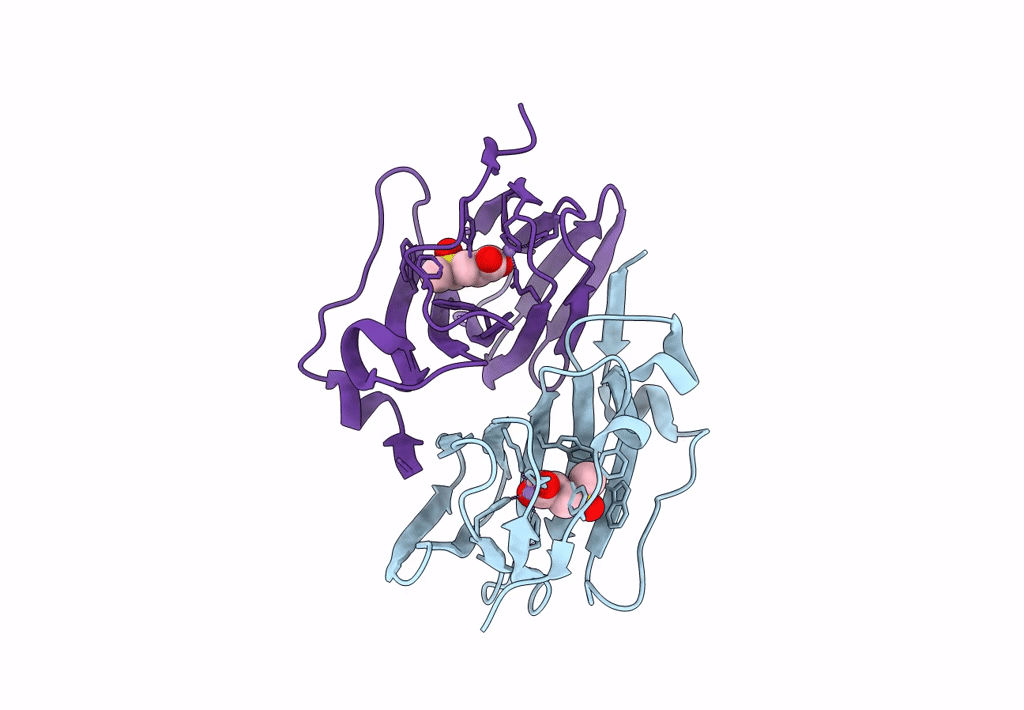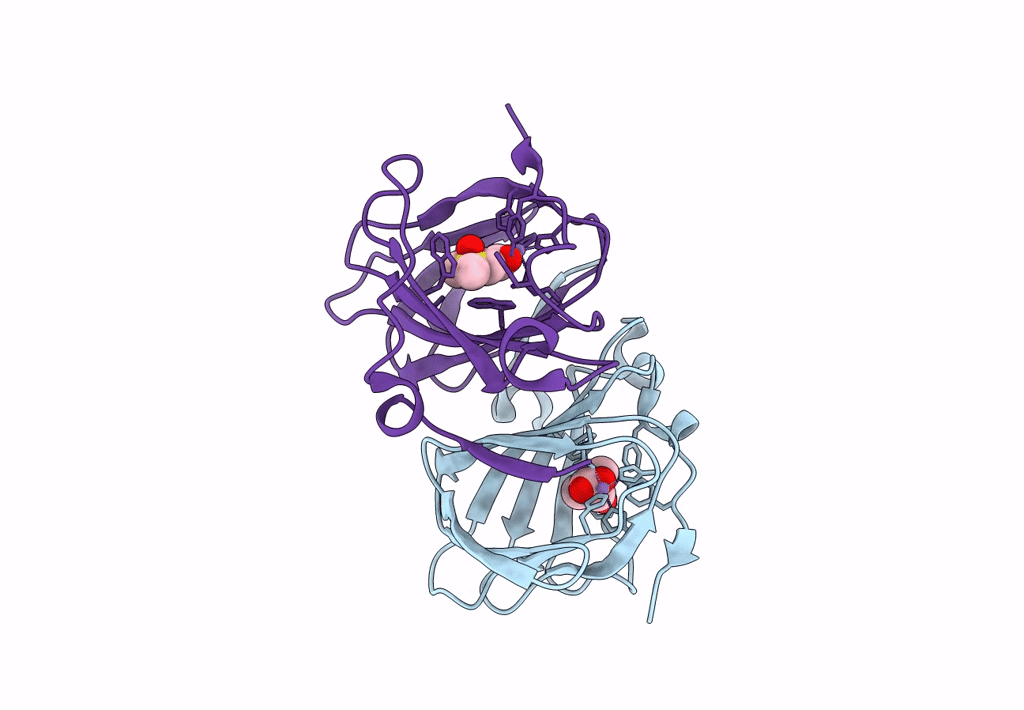 |
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-10-04 Classification: LYASE Ligands: MN, LNI |
 |
Organism: Roseovarius nubinhibens ism
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-01-25 Classification: TRANSPORT PROTEIN Ligands: DQY |
 |
Organism: Roseovarius indicus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Roseovarius indicus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: SAM, GOL, PO4 |
 |
Organism: Roseovarius indicus
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2022-04-20 Classification: TRANSFERASE Ligands: SAH, MET, PO4 |

