Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-01-31 Classification: IMMUNE SYSTEM |
 |
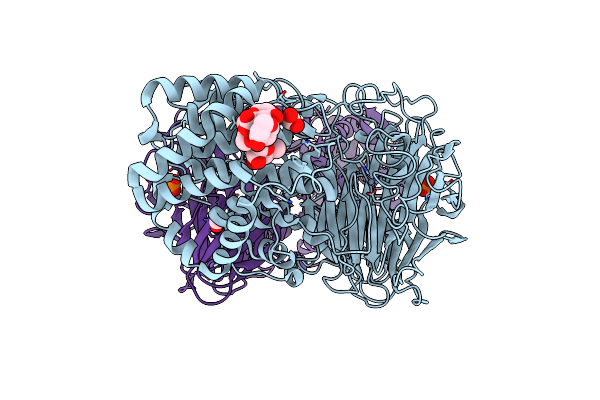
Structure Of The Active Form Of Human Origin Recognition Complex Atpase Motor Module, Complex Subunits 1, 4, 5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2017-02-08 Classification: DNA BINDING PROTEIN Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:6.00 Å Release Date: 2017-02-08 Classification: HYDROLASE |
 |
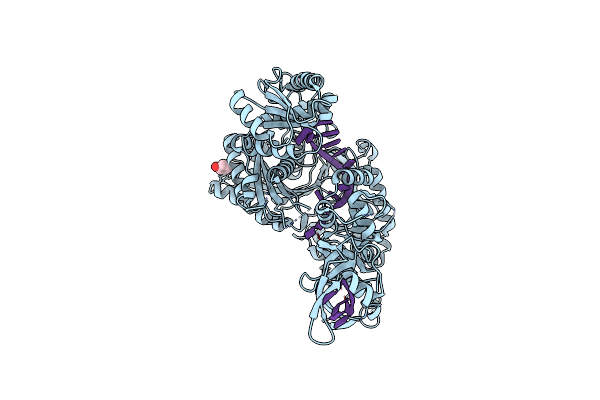
Structure Of The Active Form Of Human Origin Recognition Complex And Its Atpase Motor Module
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2017-02-08 Classification: REPLICATION Ligands: ATP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2012-05-30 Classification: Hydrolase/RNA Ligands: IPH |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-01-22 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2008-01-22 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-01-22 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2008-01-22 Classification: MEMBRANE PROTEIN |
 |
The X-Ray Structure Of A Bak Homodimer Reveals An Inhibitory Zinc Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2007-01-23 Classification: APOPTOSIS Ligands: ZN |
 |
The X-Ray Structure Of A Bak Homodimer Reveals An Inhibitory Zinc Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2006-12-26 Classification: APOPTOSIS Ligands: ZN |
 |
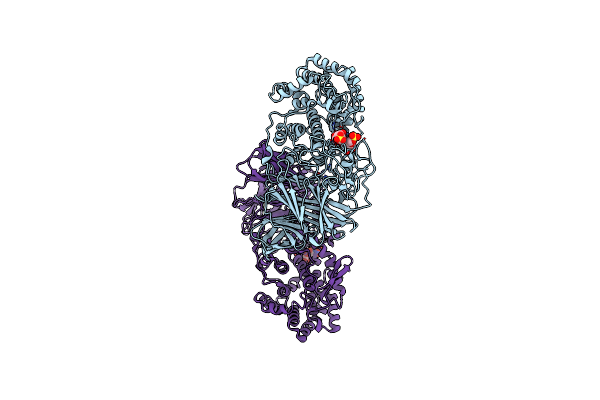
Organism: Pedobacter heparinus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2006-04-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN, PO4, FMT |
 |
Organism: Pedobacter heparinus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-04-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
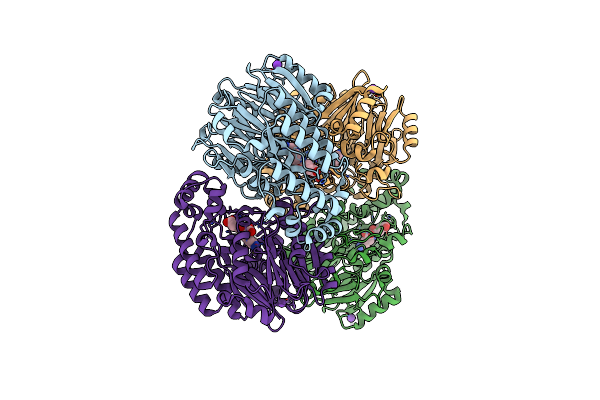
Crystal Structure Of Ureidoglycolate Hydrolase (Alla) From Escherichia Coli O157:H7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2005-10-18 Classification: HYDROLASE Ligands: GLV |
 |
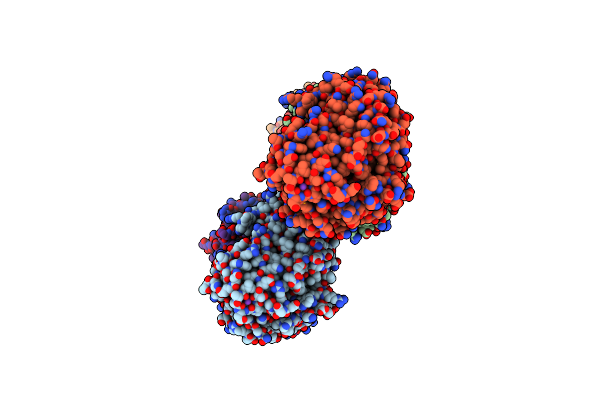
Crystal Structure Of N-Succinylarginine Dihydrolase, Astb, Bound To Substrate And Product, An Enzyme From The Arginine Catabolic Pathway Of Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2005-03-22 Classification: HYDROLASE Ligands: K, SUO |
 |
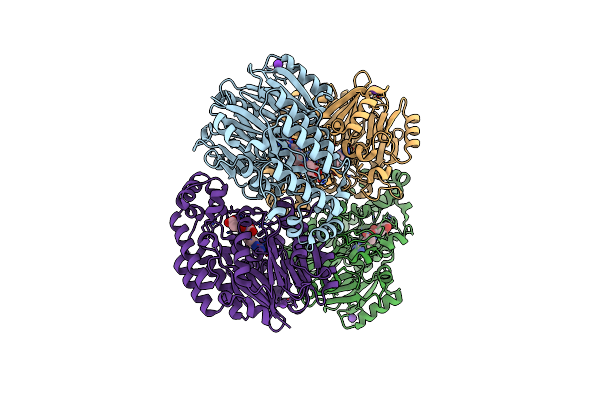
Crystal Structure Of N-Succinylarginine Dihydrolase, Astb, Bound To Substrate And Product, An Enzyme From The Arginine Catabolic Pathway Of Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-02-15 Classification: HYDROLASE Ligands: K |
 |
Crystal Structure Of N-Succinylarginine Dihydrolase, Astb, Bound To Substrate And Product, An Enzyme From The Arginine Catabolic Pathway Of Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-02-15 Classification: HYDROLASE Ligands: K, SUG |
 |
Crystal Structure Of Dodecameric Fmn-Dependent Ubix-Like Decarboxylase From Escherichia Coli O157:H7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-10-26 Classification: LYASE Ligands: FMN |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-07-06 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-05-20 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |