Search Count: 14
 |
Cryo-Em Structure Of Arabidopsis Thaliana Met1 (Aa:621-1534) In Complex With Hemimethylated Dna Analog
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: STRUCTURAL PROTEIN/DNA Ligands: SAH, ZN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-05-04 Classification: HYDROLASE |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: GOL, PGO |
 |
Crystal Structure Of Porphyromonas Endodontalis Dpp11 In Complex With Dipeptide Arg-Asp
Organism: Porphyromonas endodontalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: ARG, ASP, EDO, NA |
 |
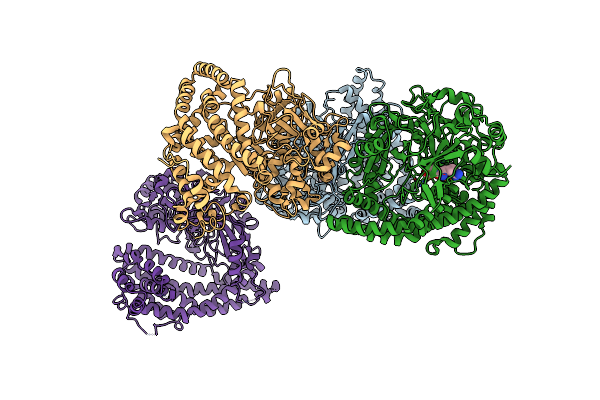
Crystal Structure Of Porphyromonas Endodontalis Dpp11 In Complex With Dipeptide Arg-Glu
Organism: Porphyromonas endodontalis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: ARG, GLU, CL |
 |
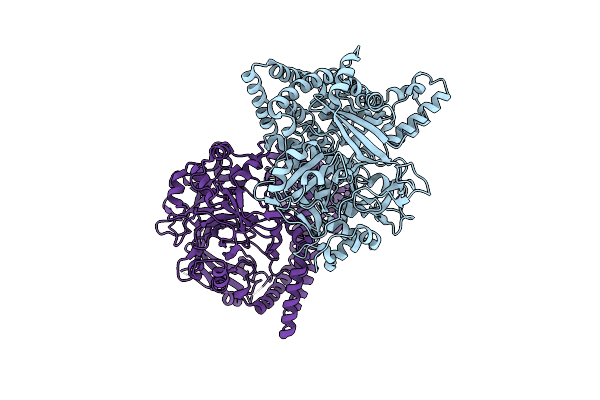
Crystal Structure Of Flavobacterium Psychrophilum Dpp11 In Complex With Dipeptide Arg-Asp
Organism: Flavobacterium psychrophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: ARG, ASP, CL, NA |
 |
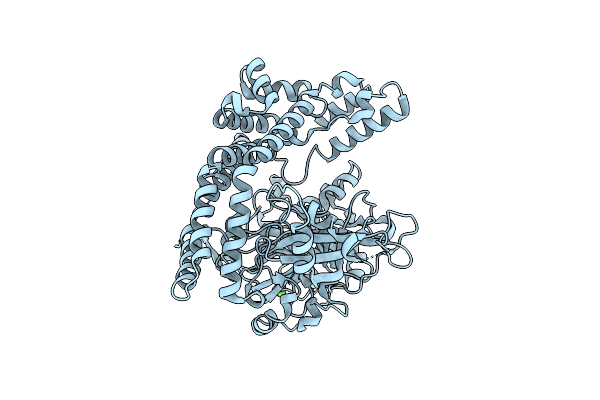
Organism: Porphyromonas endodontalis (strain atcc 35406 / bcrc 14492 / jcm 8526 / hg 370)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: CL |
 |
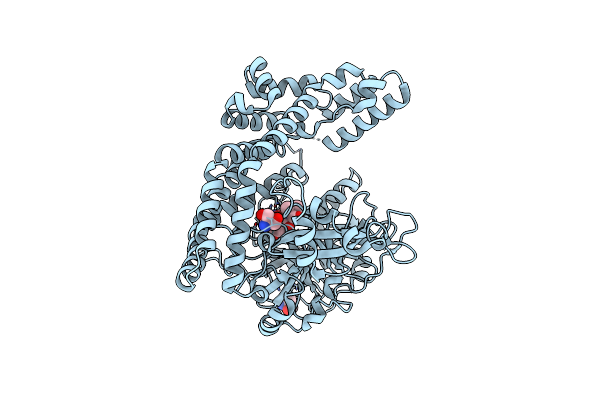
Crystal Structure Of Porphyromonas Endodontalis Dpp11 In Alternate Conformation
Organism: Porphyromonas endodontalis (strain atcc 35406 / bcrc 14492 / jcm 8526 / hg 370)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: CA, CL |
 |
Crystal Structure Of Porphyromonas Endodontalis Dpp11 In Complex With Substrate
Organism: Porphyromonas endodontalis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: TRS |
 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: NI, SO4 |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: K, CL, NA, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: CL, NA |
 |
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: CL |

