Search Count: 1,477
 |
High Resolution Structure Of The Thermophilic 60S Ribosomal Subunit Of Chaetomium Thermophilum
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, SPM, MG, K, ZN |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING |
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING Ligands: GDP, MG |
 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
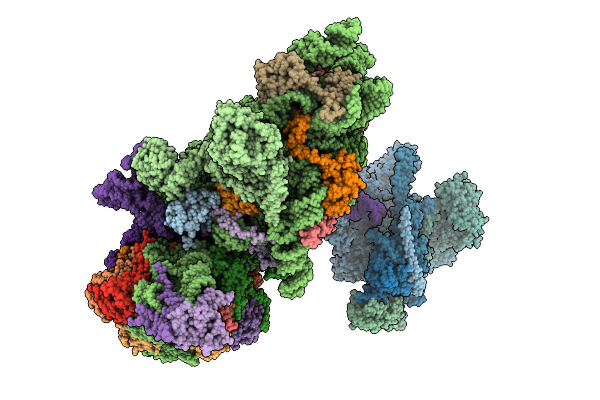
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
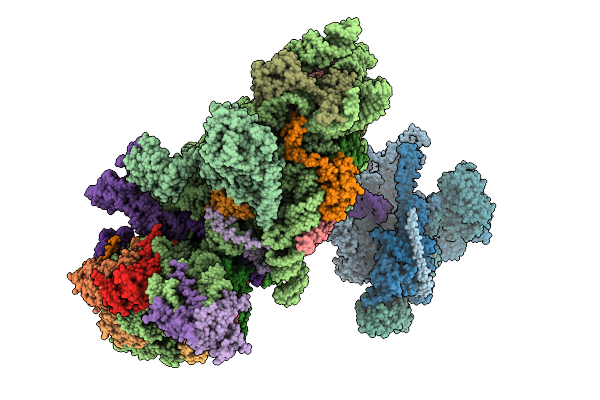
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
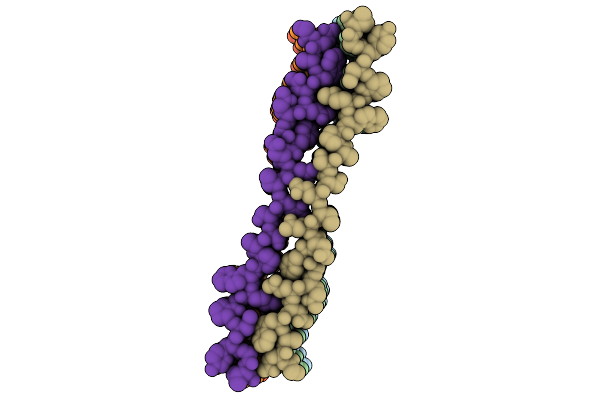 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
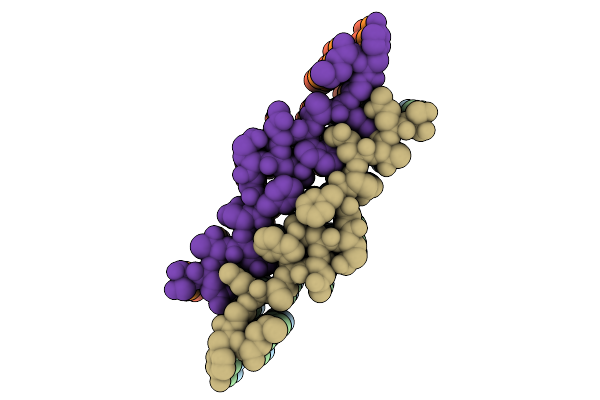 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
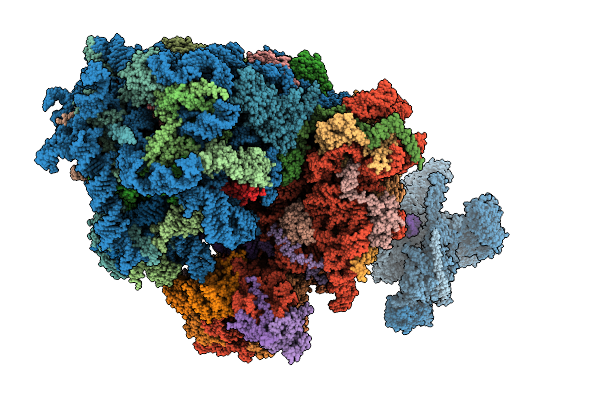 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
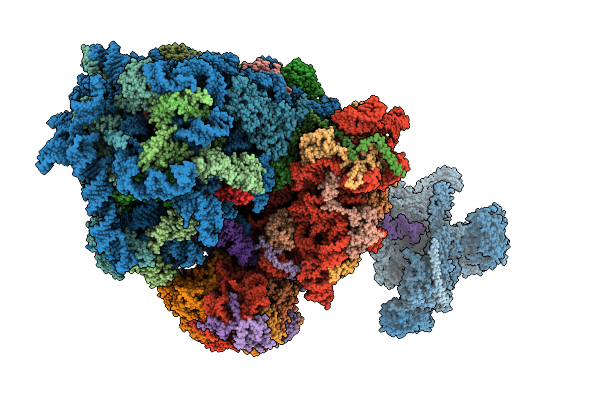 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
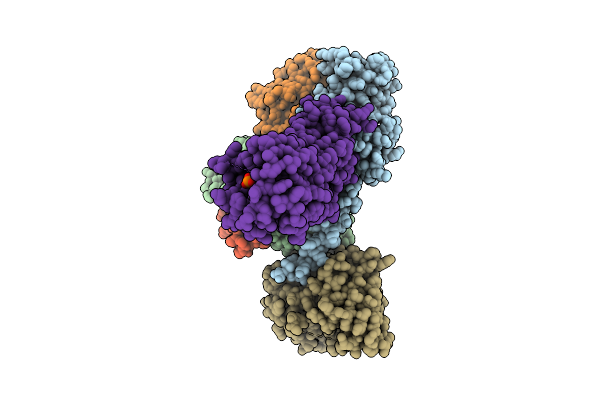 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1L69 |
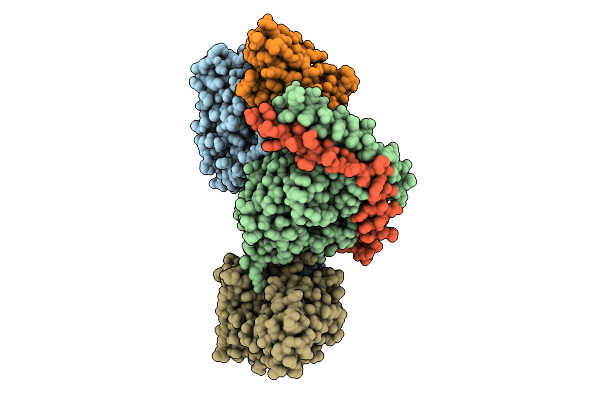 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: S1P |
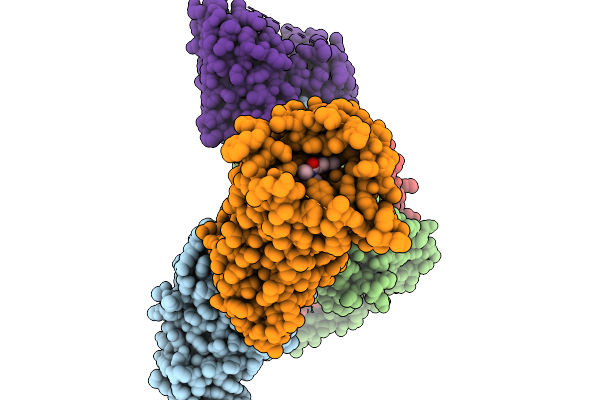 |
Cryo-Em Structure Of Human Kappa Opioid Receptor - G Protein Signaling Complex Bound With Nalfurafine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: IVB |
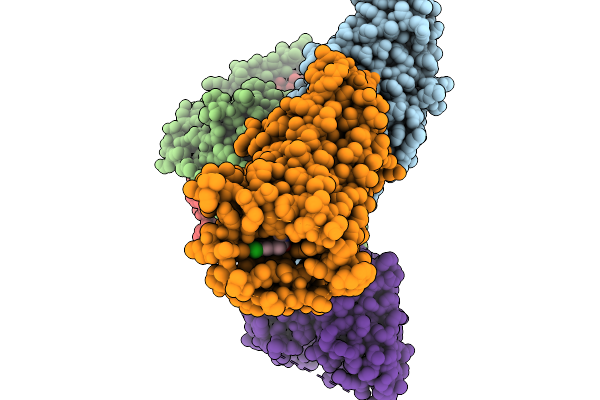 |
Cryo-Em Structure Of Human Kappa Opioid Receptor -G Protein Signaling Complex Bound With U-50488H
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: A1LWX |
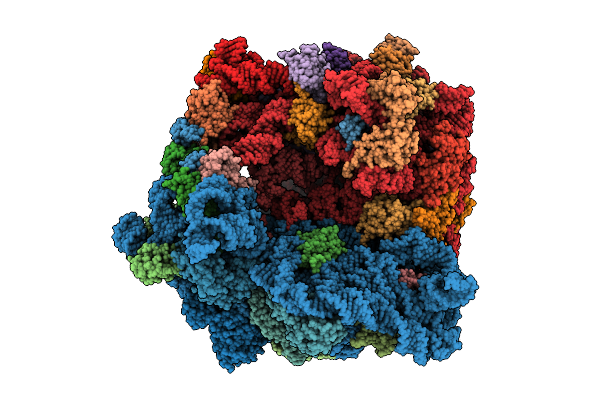 |
Structure Of The Bacterial Ribosome Without Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
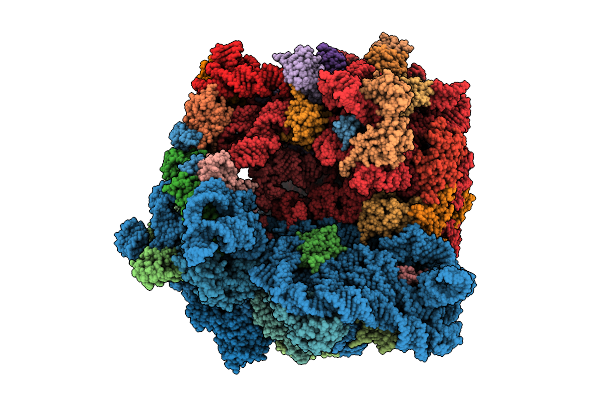 |
Structure Of The Bacterial Ribosome With Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
Organism: Synthetic construct, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE/RNA Ligands: ZN, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: GOL, ZN |
 |
Cryo-Em Structure Of Esacas9_Nng-Guide Rna-Target Dna Complex In An Interrogation State
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |

