Search Count: 23
 |
Crystallographic Structure Of Ruminococcus Champanellensis Xylanase (Rcxyn30A)
Organism: Ruminococcus champanellensis 18p13 = jcm 17042
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, PEG, NA, F, PO4 |
 |
Organism: Bacillus licheniformis dsm 13 = atcc 14580
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: TRS, GOL, CL, EDO, NA, CA |
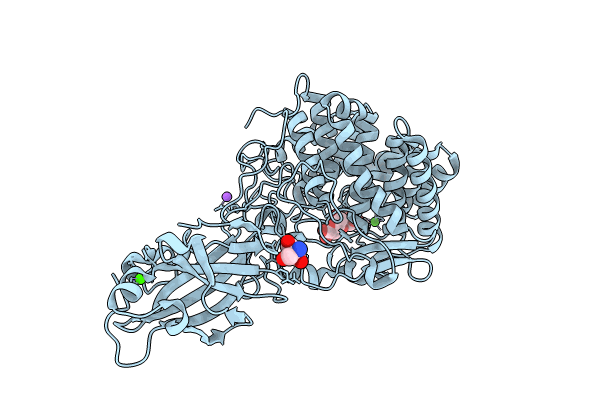 |
Crystal Structure Of Blcel9A From Glycoside Hydrolase Family 9 In Complex With Cellotriose
Organism: Bacillus licheniformis dsm 13 = atcc 14580
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: TRS, CA, NA, CL |
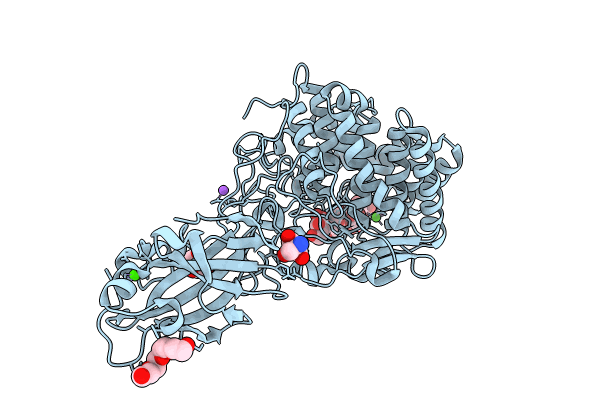 |
Crystal Structure Of Blcel9A From Glycoside Hydrolase Family 9 In Complex With Cellohexaose
Organism: Bacillus licheniformis dsm 13 = atcc 14580
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: TRS, PG4, BGC, CA, CL, NA |
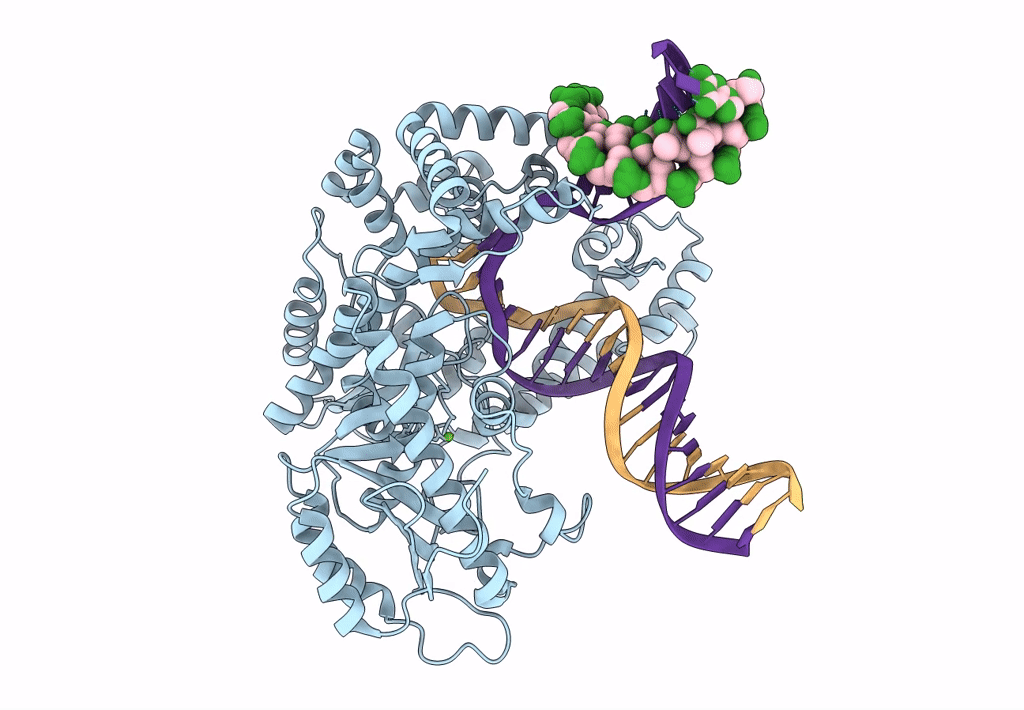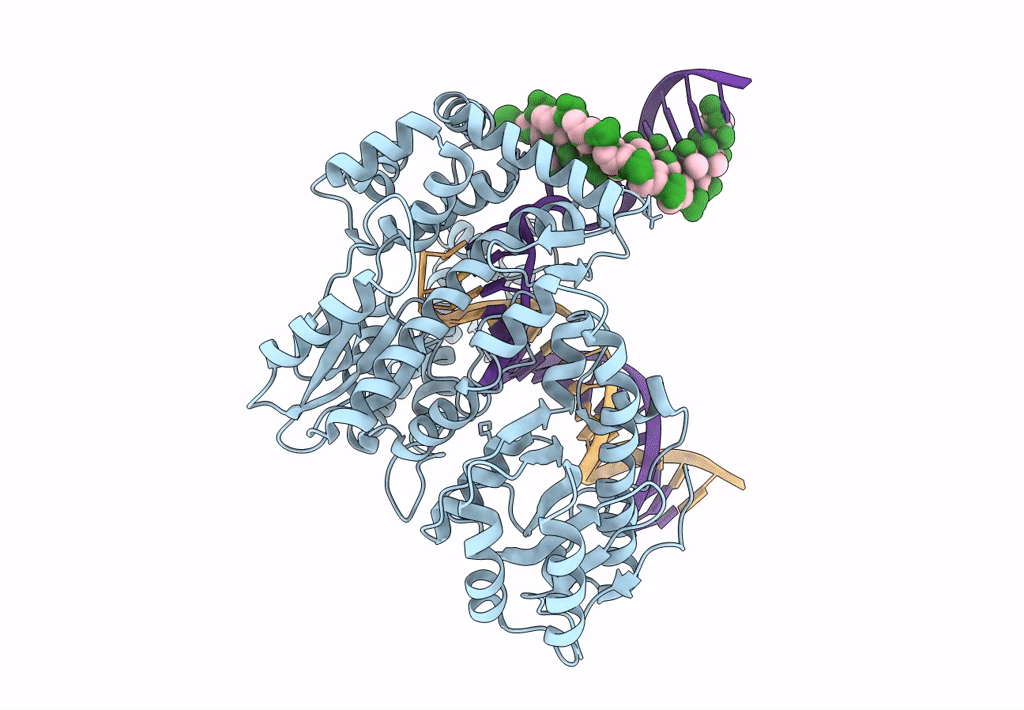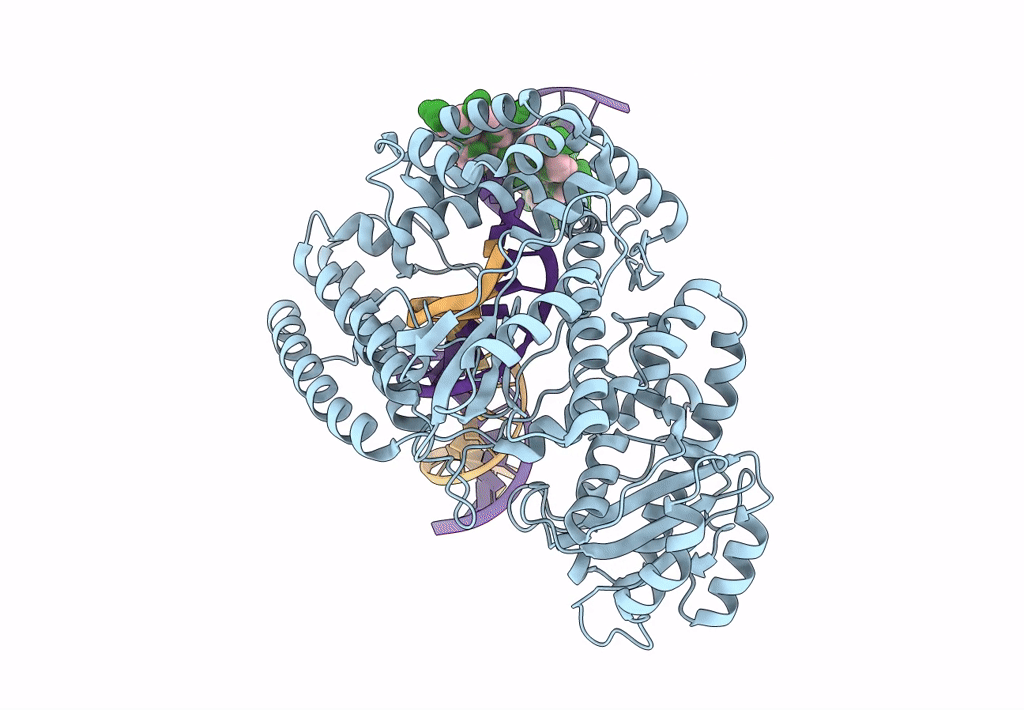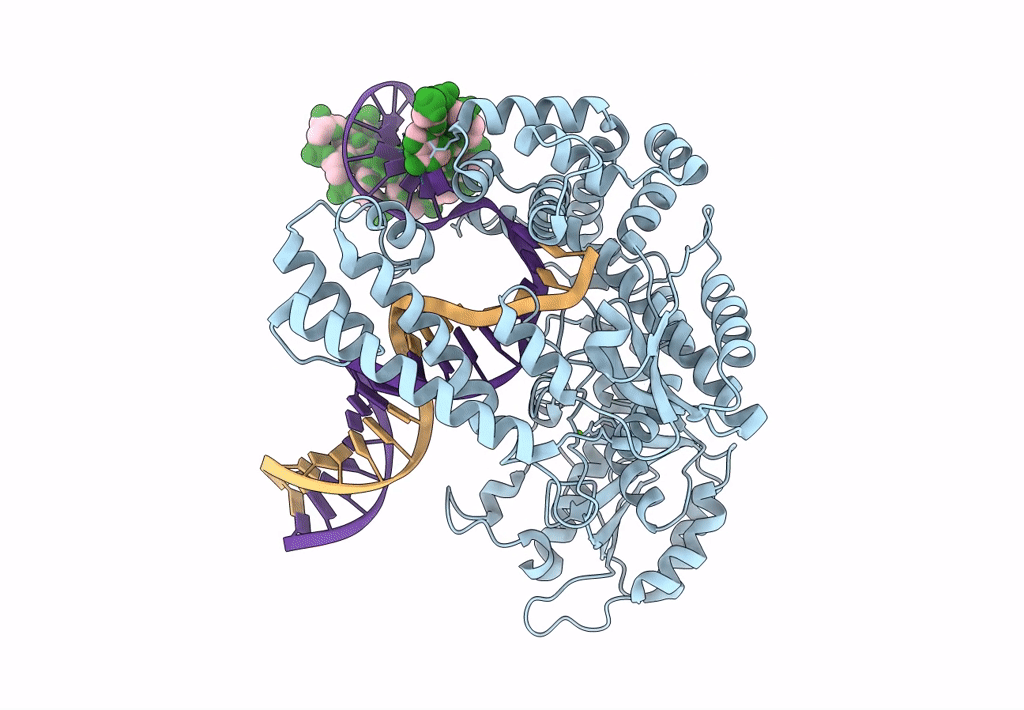 |
Organism: Escherichia coli 'bl21-gold(de3)plyss ag', Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: DNA BINDING PROTEIN Ligands: MG |
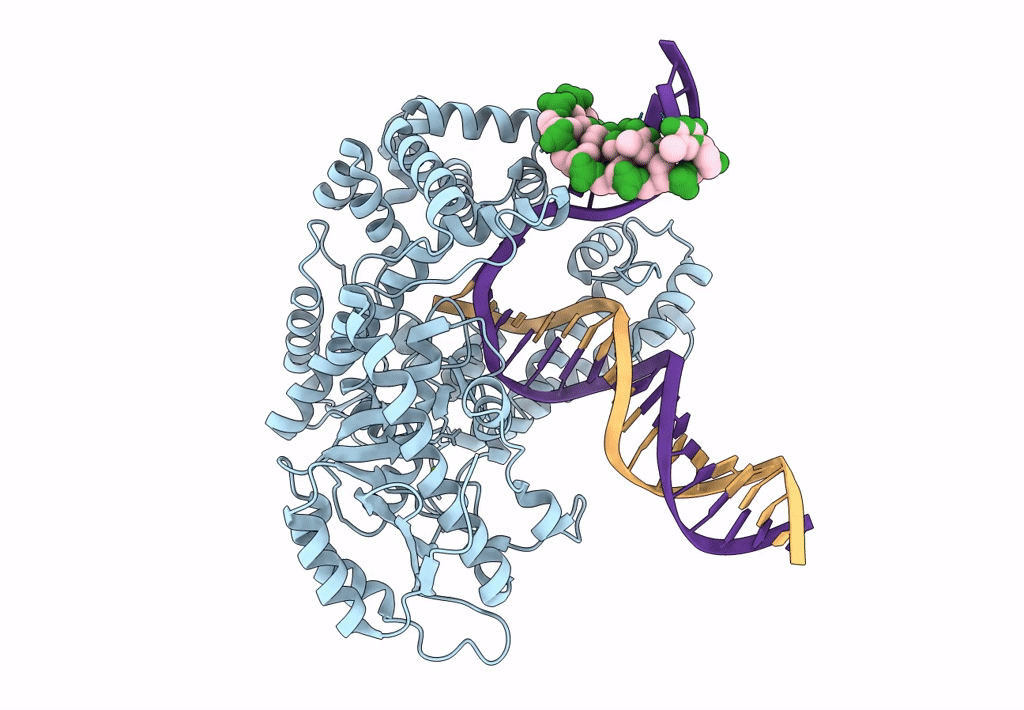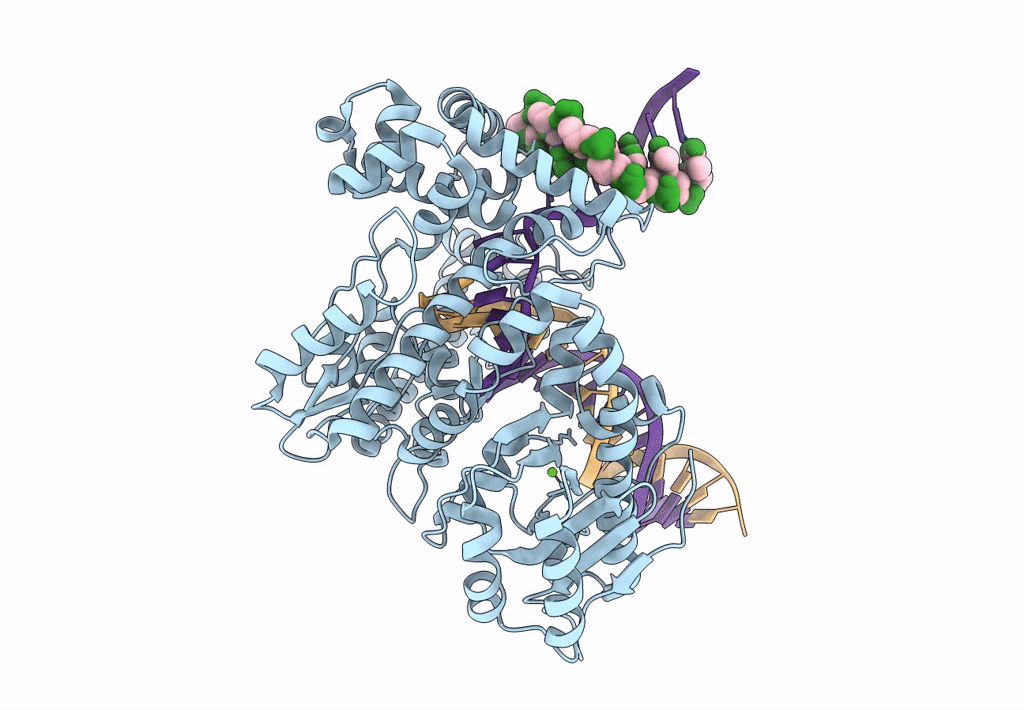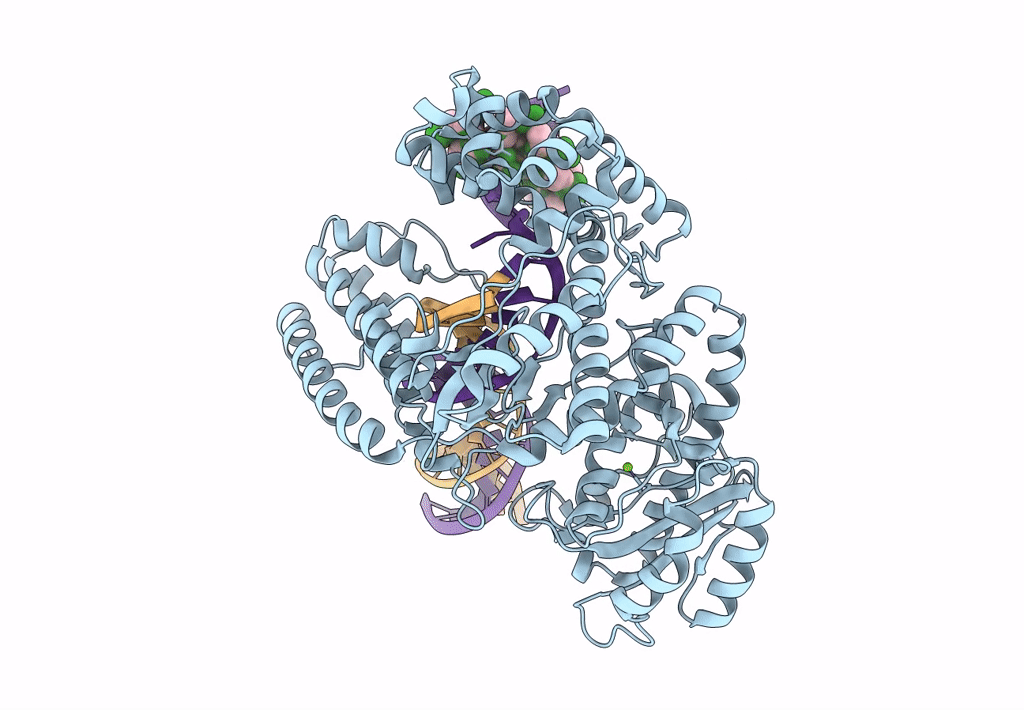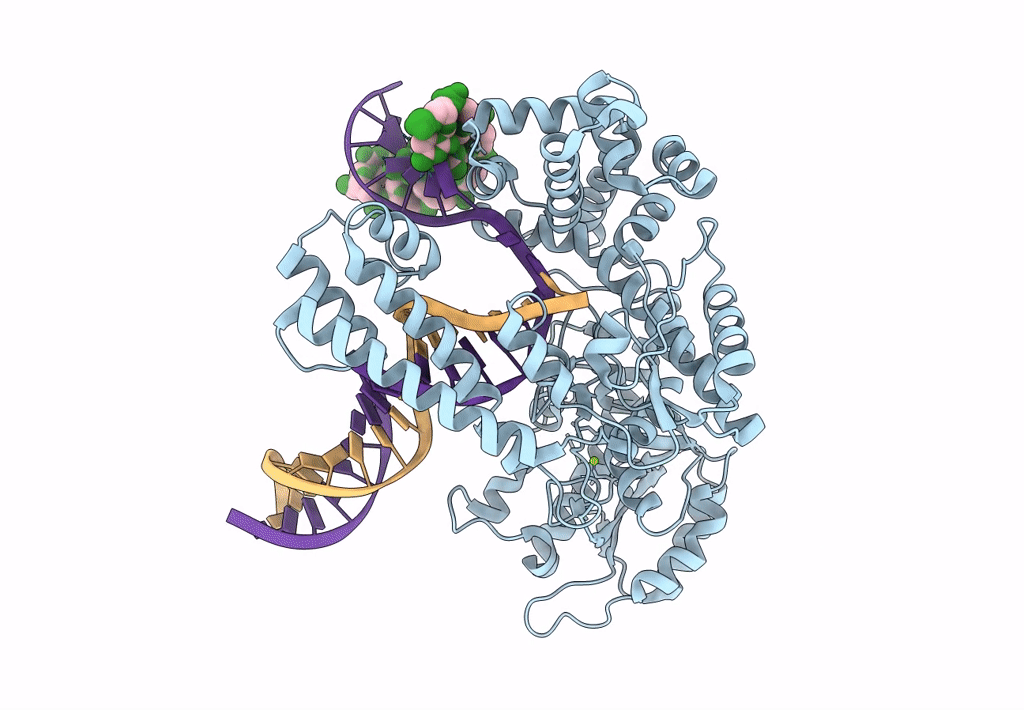 |
Organism: Escherichia coli 'bl21-gold(de3)plyss ag', Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: DNA BINDING PROTEIN Ligands: MG |
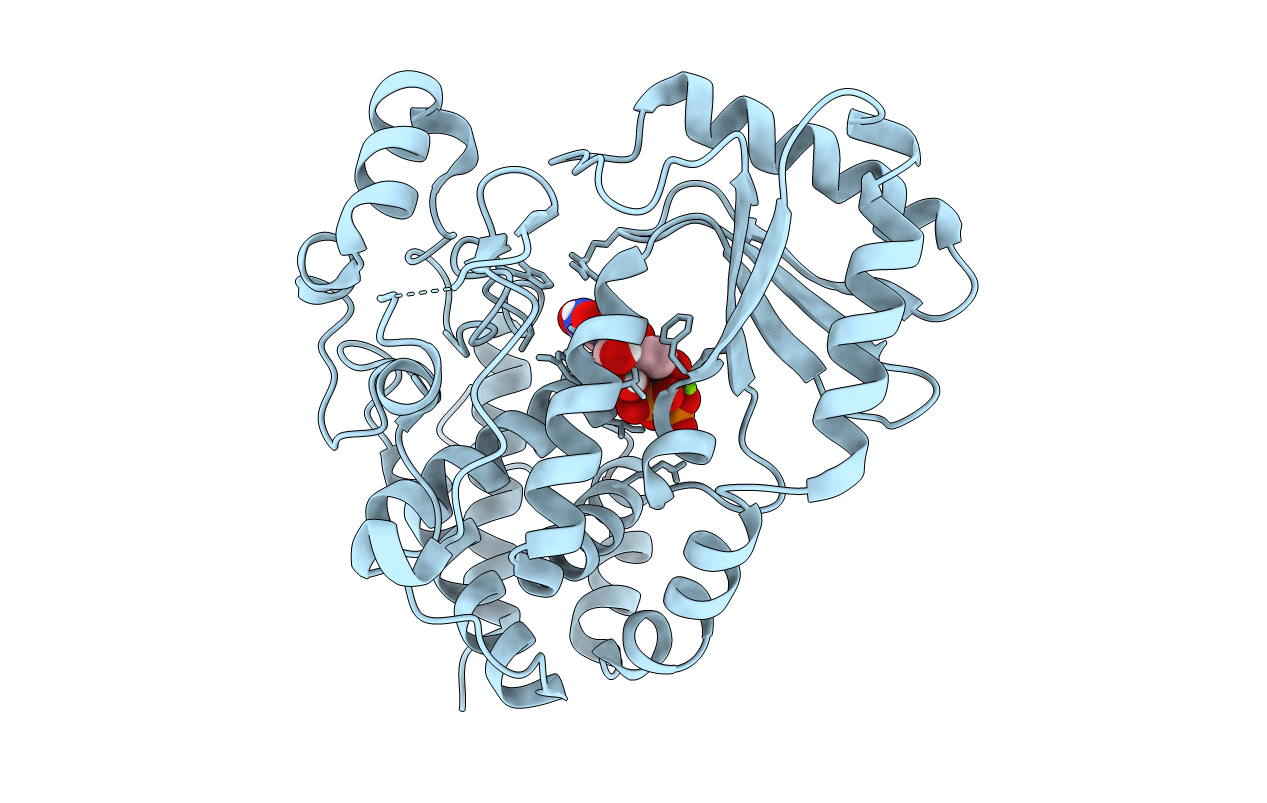 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: 2KH, MG |
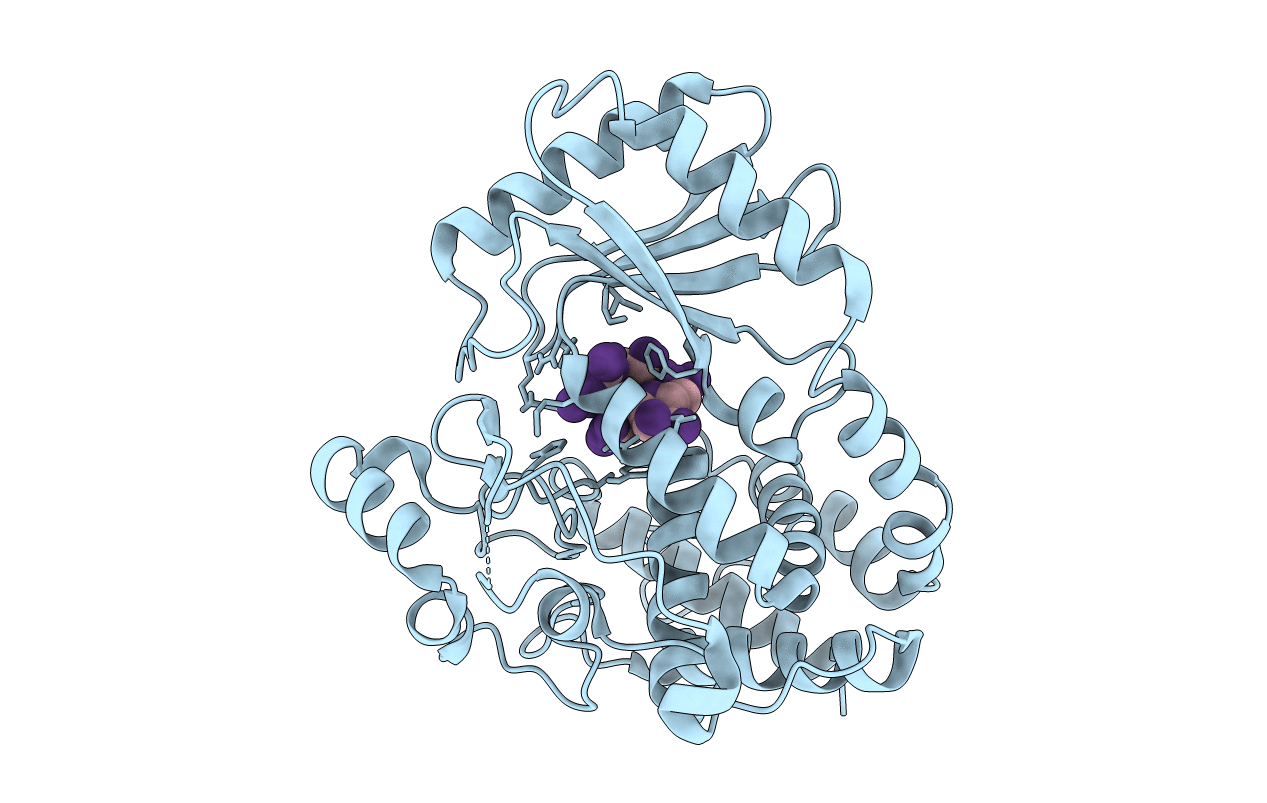 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TRANSFERASE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: MG |
 |
Crystal Structure Of The Type Ii Dehydroquinase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2014-07-23 Classification: LYASE |
 |
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-06-04 Classification: ISOMERASE Ligands: GOL, TCE |
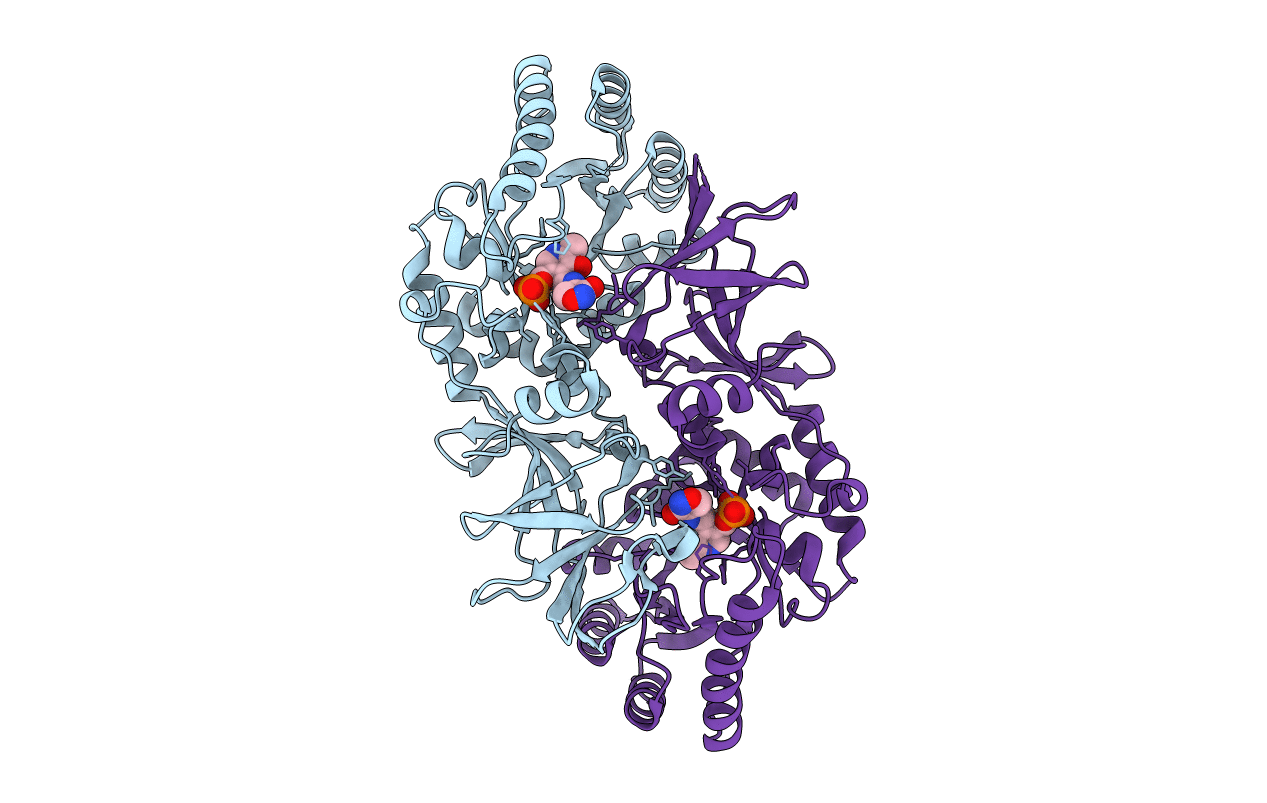 |
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-06-04 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: DCS |
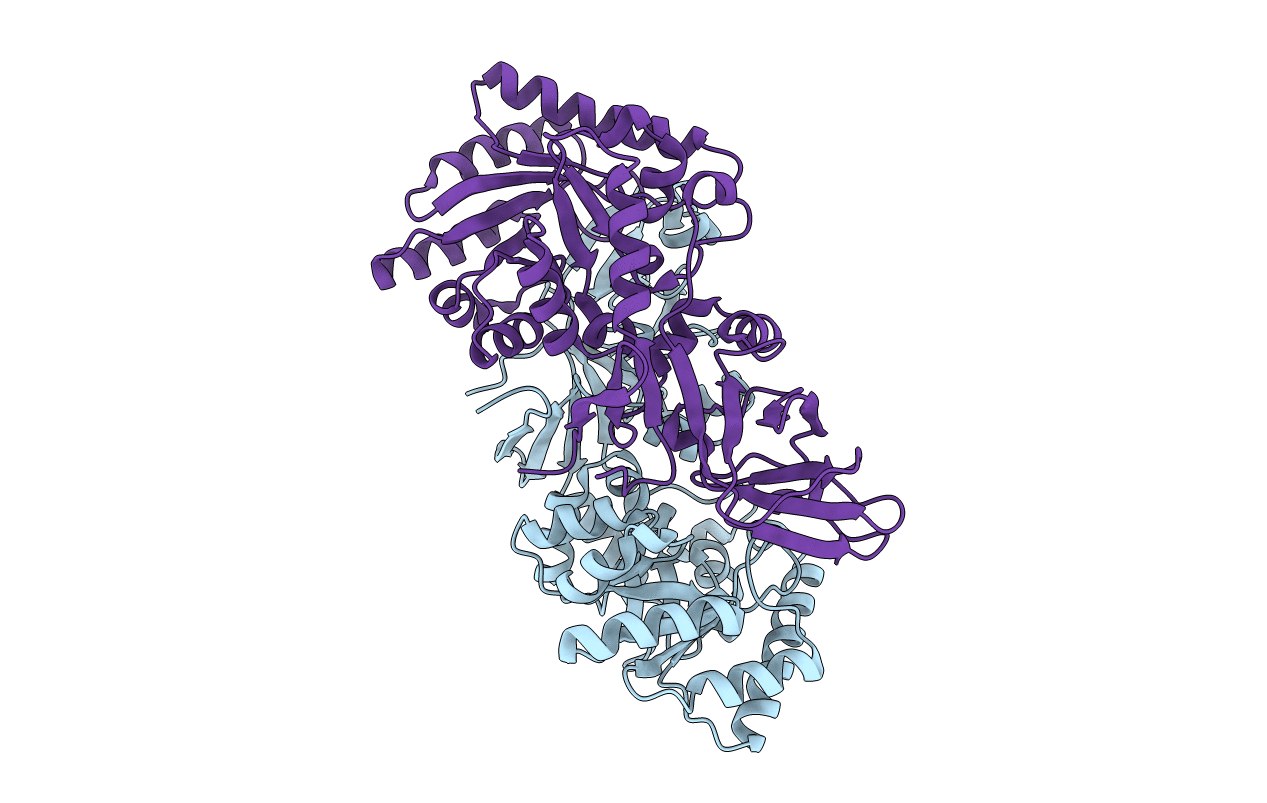 |
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-06-04 Classification: ISOMERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-10-26 Classification: HYDROLASE Ligands: EDO, PEG, ZN, PO4, DMS, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-26 Classification: Hydrolase/Inhibitor Ligands: ZN, EDO, DMS, PEG, PO4, JN4, EPE |
 |
Crystal Structure Of The Catalytic Domain Of Pde4D2 Complexed With Compound 10D
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-10-26 Classification: Hydrolase/Hydrolase Inhibitor Ligands: ZN, J25, EDO, DMS, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2011-10-26 Classification: Hydrolase/Hydrolase Inhibitor Ligands: ZN, EDO, JN8, DMS, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-10-26 Classification: Hydrolase/Hydrolase Inhibitor Ligands: EDO, ZN, PEG, JN7, PO4, DMS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ZN, ME7 |

