Search Count: 25
 |
4-Allyl Syringol Oxidase From Streptomyces Cavernae: Complex With Propanol Syringol
Organism: Streptomyces cavernae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: QBF, FAD |
 |
Organism: Streptomyces cavernae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: FAD, EOL, P6G, P4G, PEG |
 |
4-Allyl Syringol Oxidase From Streptomyces Cavernae: Complex With Vanillyl Alcohol
Organism: Streptomyces cavernae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: FAD, V55 |
 |
Vanillyl-Alcohol Dehydrogenase From Marinicaulis Flavus: P151L Mutant Bound To Eugenol
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, 1PE, PE4, EOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
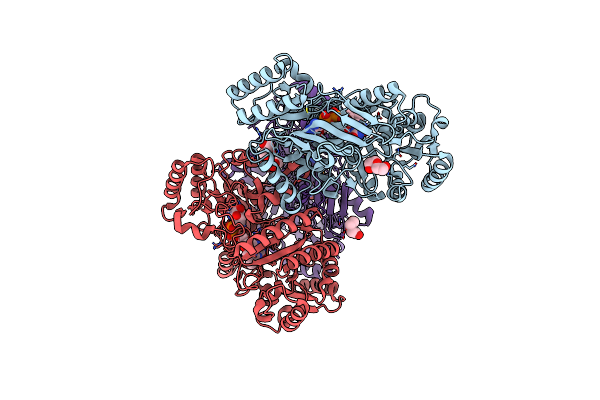
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, GOL |
 |
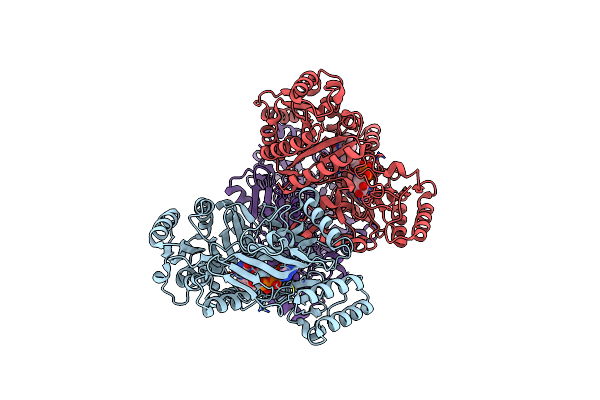
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PEG |
 |
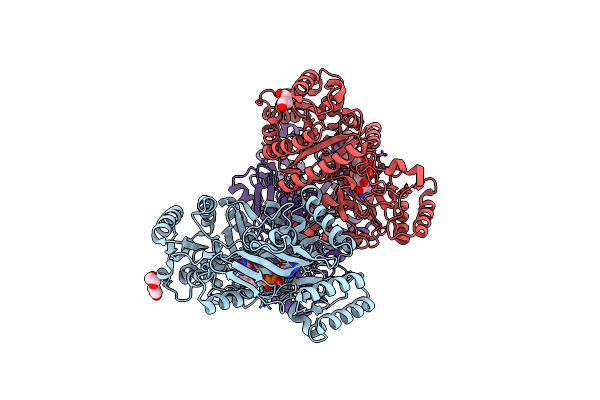
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
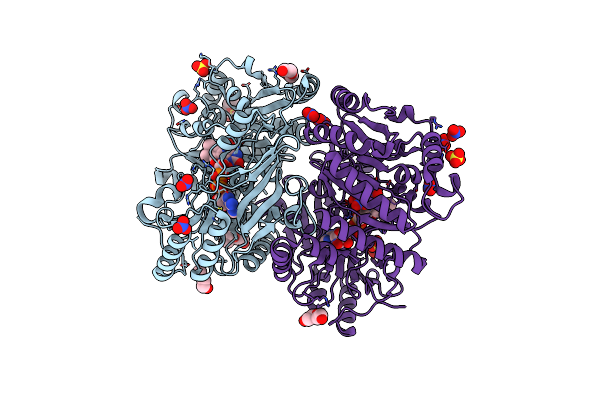
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, SO4 |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PEG |
 |
Vanillyl Alcohol Oxidase From Novosphingobium Sp In Complex With Vanillyl Alcohol
Organism: Novosphingobium sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: FAD, GOL, V55 |
 |
Vanillyl Alcohol Oxidase From Novosphingobium Sp: T181D Mutant In Complex With Vanillin
Organism: Novosphingobium sp. 01wb02.4-10
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-15 Classification: FLAVOPROTEIN Ligands: FAD, V55, GOL, PEG |
 |
Cryo-Em Structure Of Styrene Oxide Isomerase Bound To Benzylamine Inhibitor
Organism: Pseudomonas sp. vlb120, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN Ligands: ABN, HEM |
 |
Organism: Pseudomonas sp. vlb120, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN Ligands: HEM |
 |
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070 In Complex With Udp
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: UDP, GOL, NA, EDO |
 |
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070 In Complex With Udp-Glucose
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: UPG, EDO, PEG, SCN, MG |
 |
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: PEG, EDO, SCN, K |
 |
Crystal Structure Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Fad
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Variovorax Paradoxus Indole Monooxygenase (Vpinda1) In Complex With Indole
Organism: Variovorax paradoxus eps
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, IND, SO4, DMS, NA |

