Search Count: 6
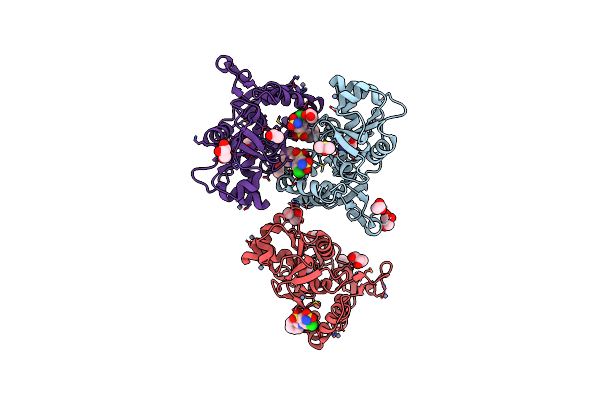 |
Crystal Structure Of The Iglur2 Ligand-Binding Core (S1S2J-N754S) In Complex With Glutamate And Cyclothiazide At 2.25 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-07-28 Classification: MEMBRANE PROTEIN Ligands: GLU, CYZ, ZN, GOL, ACT, DMS, CAC |
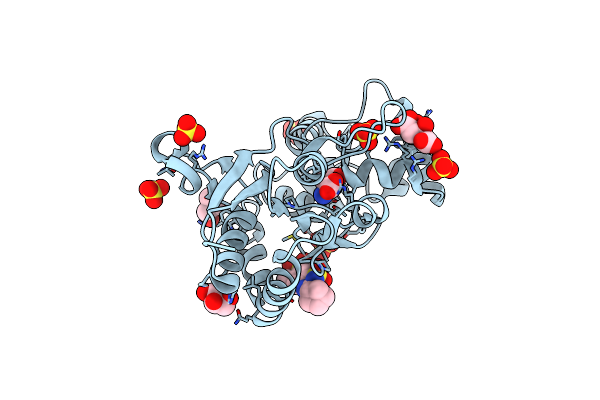 |
Crystal Structure Of The Iglur2 Ligand-Binding Core (S1S2J-N754S) In Complex With Glutamate And Ns1493 At 1.85 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-07-28 Classification: MEMBRANE PROTEIN Ligands: GLU, NS3, SO4, GOL, FLC |
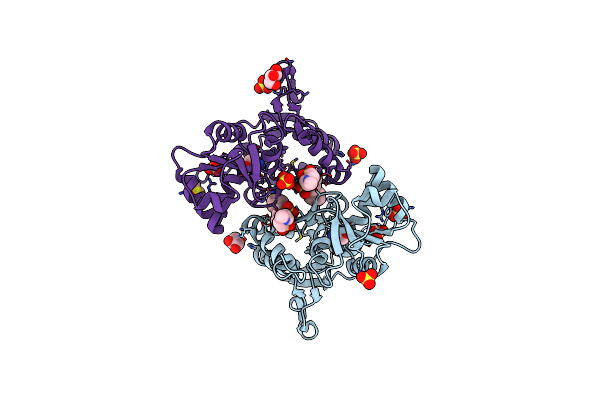 |
Crystal Structure Of The Iglur2 Ligand-Binding Core (S1S2J-N754S) In Complex With Glutamate And Ns5206 At 2.10 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-07-28 Classification: MEMBRANE PROTEIN Ligands: GLU, NS6, GOL, SO4, DMS |
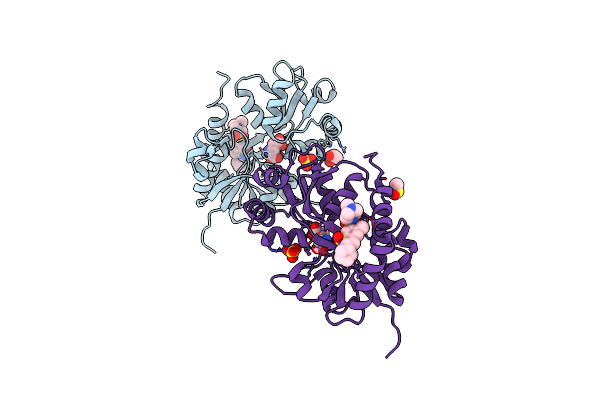 |
Crystal Structure Of The Iglur2 Ligand-Binding Core (S1S2J-N754S) In Complex With Glutamate And Ns5217 At 1.50 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2009-07-28 Classification: MEMBRANE PROTEIN Ligands: GLU, NS7, SO4, GOL, DMS |
 |
Crystal Structure Of The Ligand-Binding Core Of Iglur5 In Complex With The Antagonist (S)-Atpo At 1.85 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-07-03 Classification: MEMBRANE PROTEIN Ligands: AT1, GOL |
 |
Crystal Structure Of The Ligand-Binding Core Of Iglur5 In Complex With The Partial Agonist Domoic Acid At 2.5 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-03 Classification: MEMBRANE PROTEIN Ligands: DOQ |

