Search Count: 58
 |
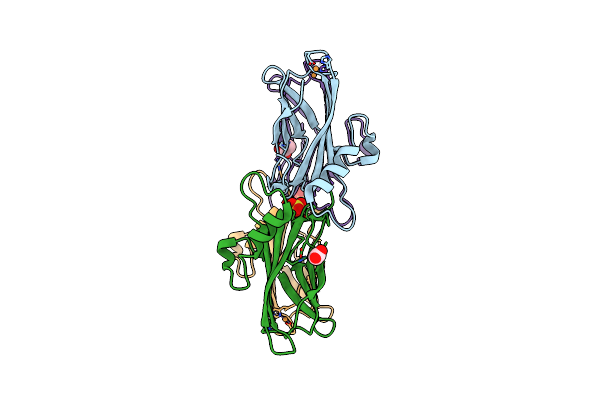
Nmr Solution Structure Of Termini-Truncated Oxidized Cytochrome C552 From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: SOLUTION NMR Release Date: 2025-07-30 Classification: ELECTRON TRANSPORT Ligands: HEC |
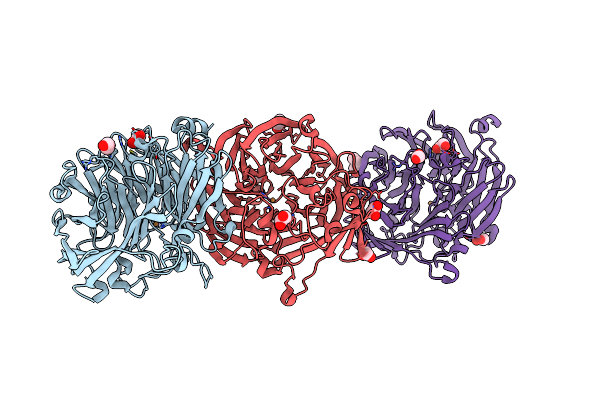 |
The Structure Of Non-Activated Thiocyanate Dehydrogenase From Pelomicrobium Methylotrophicum (Pmtcdh)
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: EDO, CL, CU, PEG |
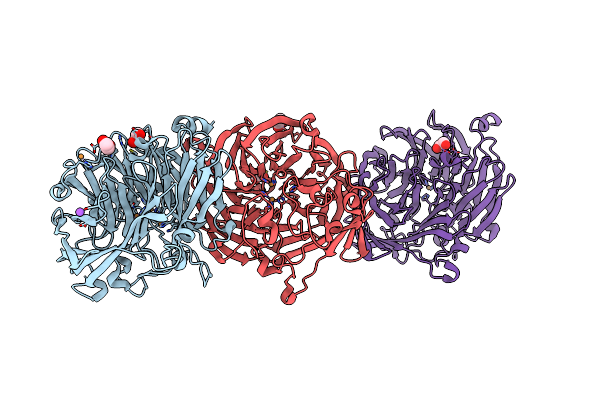 |
The Structure Of Thiocyanate Dehydrogenase From Pelomicrobium Methylotrophicum (Pmtcdh), Activated By Crystals Soaking With 1 Mm Cucl2 During 6 Months
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: EDO, CU, NA |
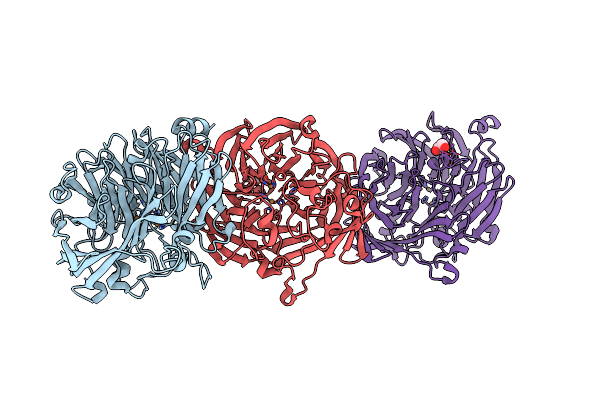 |
The Structure Of Thiocyanate Dehydrogenase From Pelomicrobium Methylotrophicum (Pmtcdh), Activated By Crystals Soaking With 1 Mm Cucl2 And Na Ascorbate During 12 Hours
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CU, EDO |
 |
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, SO4 |
 |
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: PEG, CU |
 |
The Structure Of The Cytochrome C546/556 From Thioalkalivibrio Paradoxus With Unusual Uv-Vis Spectral Features At Atomic Resolution
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Release Date: 2024-04-24 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
The Structure Of Non-Activated Thiocyanate Dehydrogenase Mutant With The H447Q Substitution From Pelomicrobium Methylotrophicum (Pmtcdh H447Q)
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: PEG, EDO, CU, CL |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant With The H447Q Substitution From Pelomicrobium Methylotrophicum (Pmtcdh H447Q), Activated By Crystal Soaking With 1Mm Cucl2 And 1 Mm Sodium Ascorbate
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-04-24 Classification: OXIDOREDUCTASE Ligands: EDO, CU, CL |
 |
Crystal Structure Of Octaheme Nitrite Reductase From Trichlorobacter Ammonificans In Complex With Nitrite
Organism: Trichlorobacter ammonificans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: HEC, NO, NO2, CA |
 |
Crystal Structure Of Octaheme Nitrite Reductase From Trichlorobacter Ammonificans In Space Group P63
Organism: Trichlorobacter ammonificans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: HEC, PO4, CA |
 |
Crystal Structure Of Octaheme Nitrite Reductase From Trichlorobacter Ammonificans In Space Group P21
Organism: Trichlorobacter ammonificans
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: HEC, CA, PO4 |
 |
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: SEK, CU, GOL, SE, NA |
 |
The Structure Of Thiocyanate Dehydrogenase From Pelomicrobium Methylotrophicum With Molecular Oxygen At 1.05 A Resolution
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: GOL, OXY, CU |
 |
The Structure Of Thiocyanate Dehydrogenase From Pelomicrobium Methylotrophicum In Complex With Inhibitor Thiourea At 1.10 A Resolution
Organism: Pelomicrobium methylotrophicum
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: GOL, TOU, CU |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant Form With Lys 281 Replaced By Ala From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: SO4, BO3, CU, NA |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant Form With Thr 169 Replaced By Ala From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant Form With Phe 436 Replaced By Gln From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-01-18 Classification: OXIDOREDUCTASE Ligands: PEG, CU |
 |
Solution Structure Of Oxidized Cytochrome C552 From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: SOLUTION NMR Release Date: 2021-05-05 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Carboxydothermus ferrireducens dsm 11255
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE Ligands: HEC, TRS, GOL, CA, PO4 |

