Search Count: 14
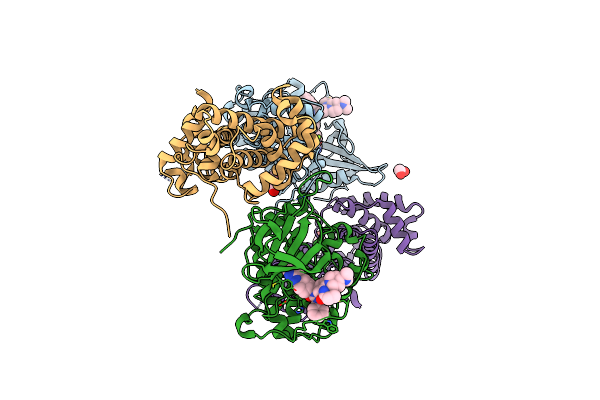 |
Crystal Structure Of Cdk12/Cyclin K In Complex With Covalent Inhibitor Yjz5118
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2025-05-07 Classification: TRANSFERASE Ligands: EDO, A1EB3, MG, PEG |
 |
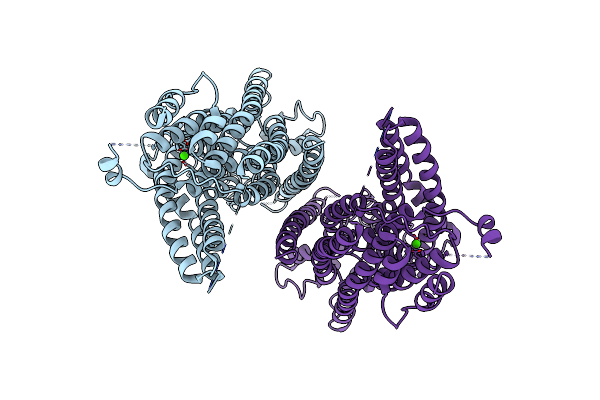
Cryo-Em Structure Of The Tmem16A Calcium-Activated Chloride Channel In Nanodisc
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of The Tmem16A Calcium-Activated Chloride Channel In Lmng
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
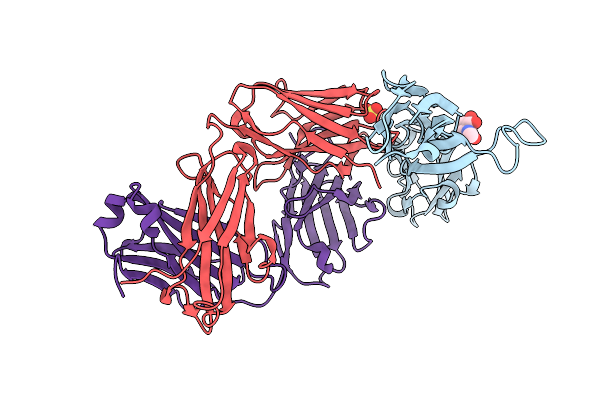
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2015-09-16 Classification: TRANSFERASE/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-10-29 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GOL, 2UG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-05-05 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: NAG, SO4 |
 |
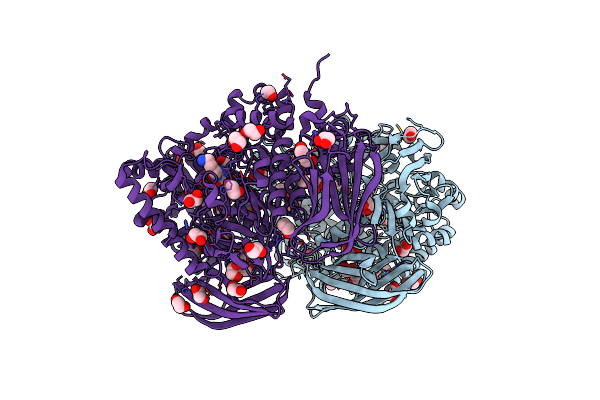
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-08 Classification: HYDROLASE Ligands: 15A, EDO, BR, CL |
 |
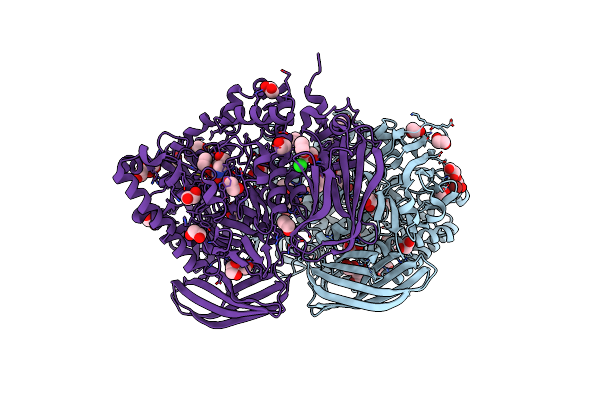
Transition-State Mimicry In Mannoside Hydrolysis: Characterisation Of Twenty Six Inhibitors And Insight Into Binding From Linear Free Energy Relationships And 3-D Structure
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-04-08 Classification: HYDROLASE Ligands: 17B, EDO, BR, CL |
 |
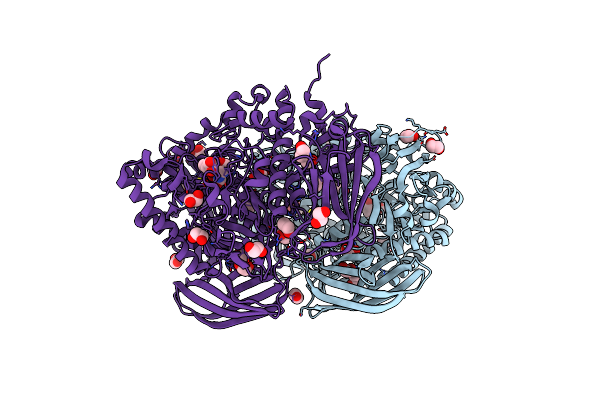
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: IFL, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MNM, EDO, BR, CL |
 |
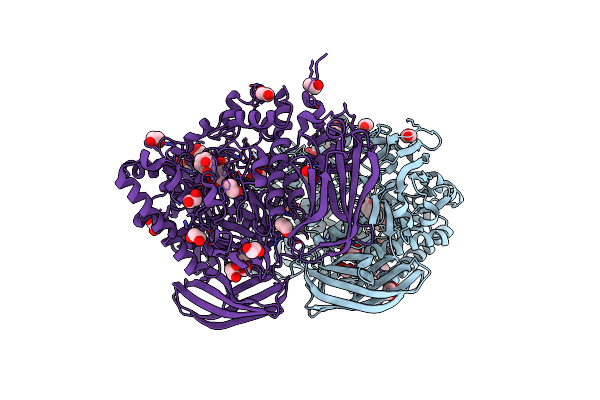
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: MVL, EDO, BR, CL |
 |
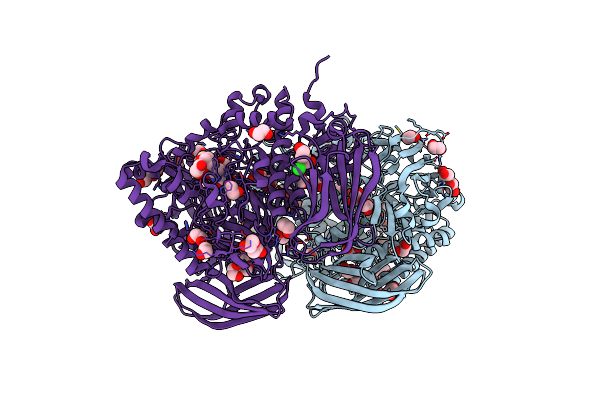
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: VBZ, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: NHV, EDO, BR, CL |
 |
Structural And Biochemical Evidence For A Boat-Like Transition State In Beta-Mannosidases
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-01 Classification: HYDROLASE Ligands: EDO, NOY, BR, CL |

