Search Count: 199
 |
Organism: Mus musculus, Monkeypox virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Mus musculus, Monkeypox virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PROTEIN BINDING |
 |
Aagab Pseudogtpase Domain In Complex With Ap-1 Clathrin Adaptor Complex Sigma 3 Subunit
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PROTEIN BINDING |
 |
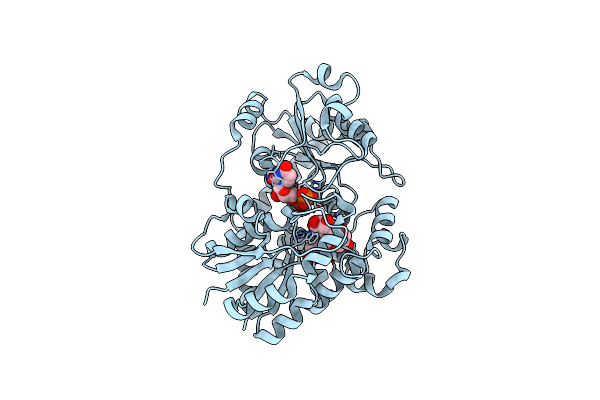
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: UDP |
 |
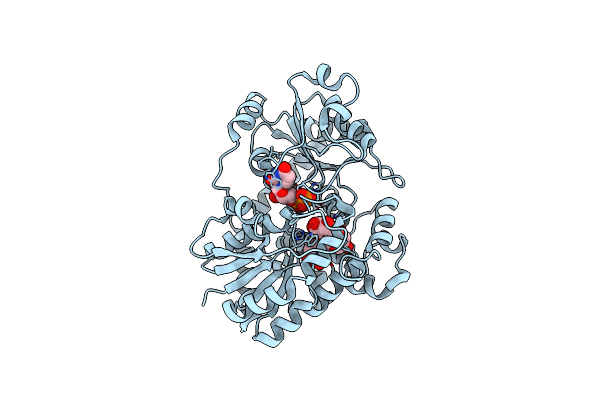
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: UDP, A1EGV |
 |
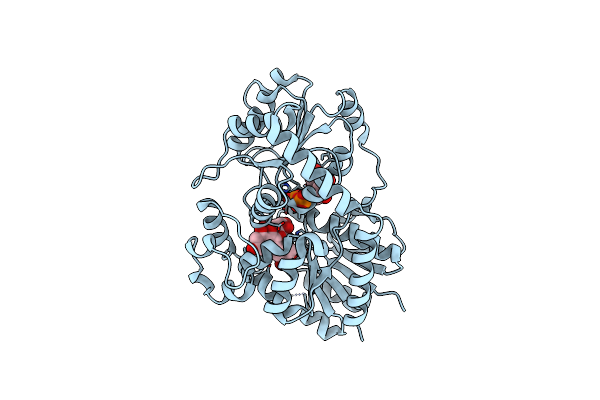
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: HW2, UDP |
 |
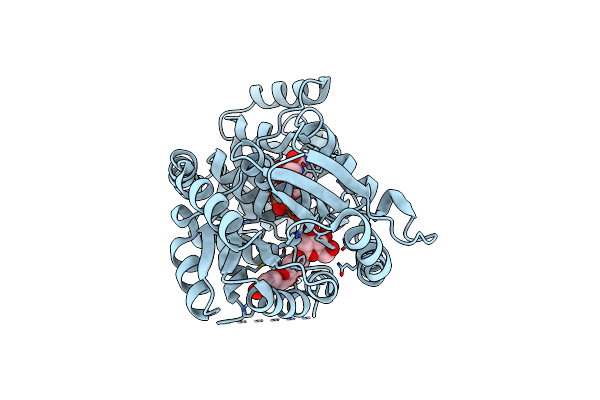
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: J6O, UDP |
 |
Organism: Petroselinum crispum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: A1EGT, UDP |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: U5P, RUT |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: A1EGW, UDP |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: A1EGX, UDP |
 |
Organism: Petroselinum crispum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: UDP |
 |
Organism: Petroselinum crispum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: PLANT PROTEIN Ligands: A1EGY, UDP |
 |
The Crystal Structure Of An Atypical N-Methyltransferasea Paomt9 In P. Amurense
Organism: Phellodendron amurense
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: SAM, A1D8J |
 |
The X-Ray Crystallographic Structure Of Oligo-1,6-Glucosidase From Paenibacillus Sp. Stb16
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2025-05-07 Classification: HYDROLASE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-23 Classification: TRANSFERASE |
 |
Organism: Escherichia fergusonii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: IMMUNE SYSTEM Ligands: ATP, MG |

