Search Count: 28
 |
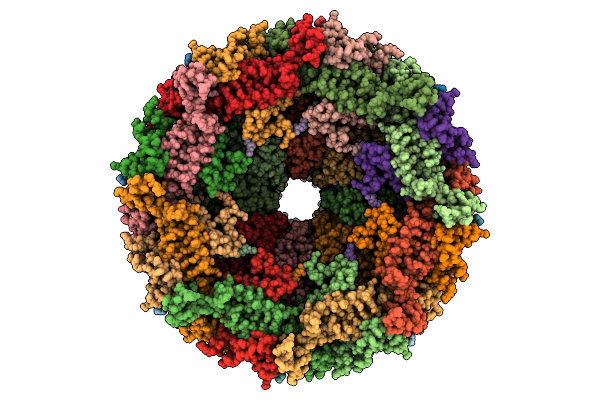
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
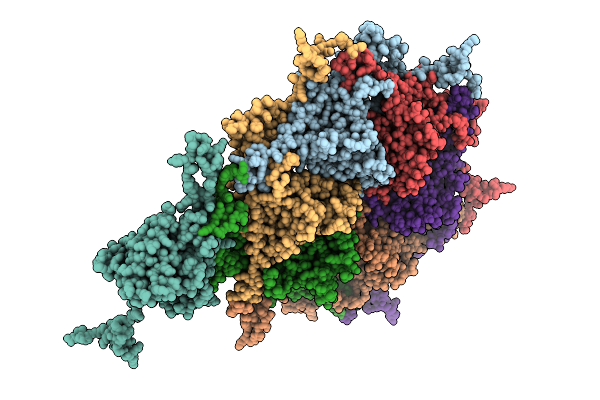
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
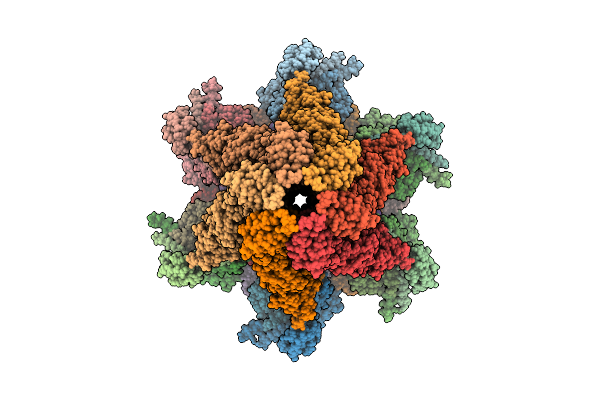
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
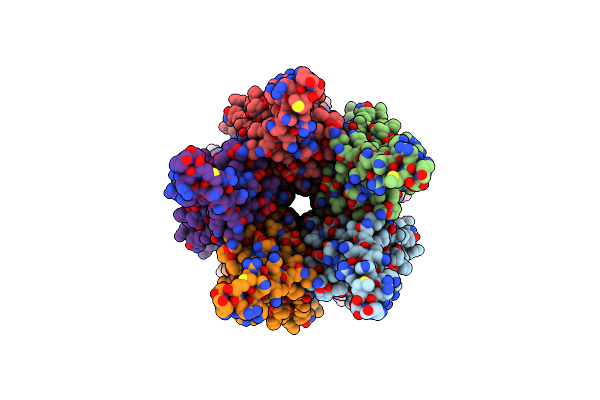
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Organism: Caenorhabditis elegans, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: TRANSPORT PROTEIN Ligands: NAG, A1D8E, BET |
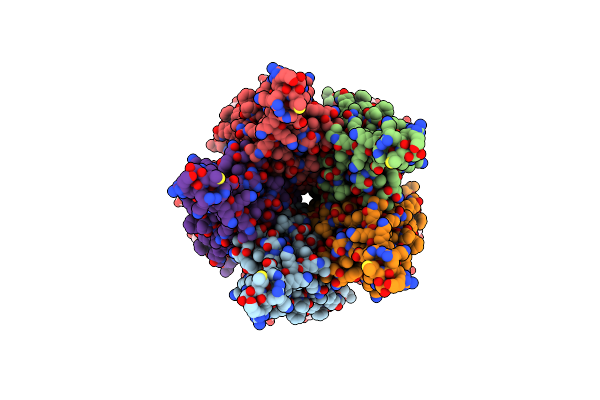 |
Organism: Caenorhabditis elegans, Shimwellia blattae
Method: ELECTRON MICROSCOPY Resolution:2.61 Å Release Date: 2024-07-31 Classification: TRANSPORT PROTEIN Ligands: NAG |
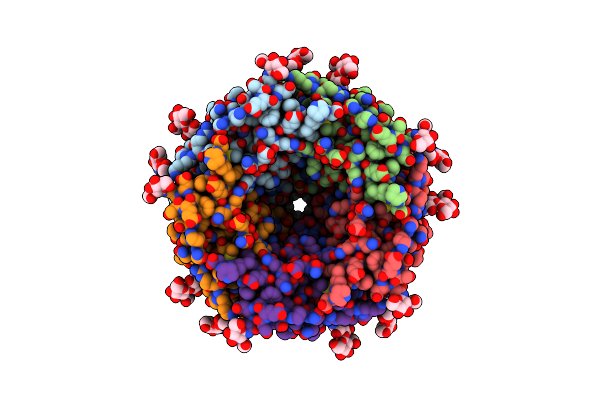 |
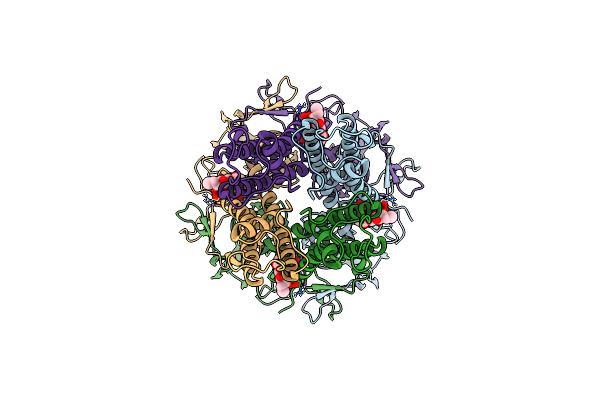
Organism: Caenorhabditis elegans, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: TRANSPORT PROTEIN Ligands: NAG, BET |
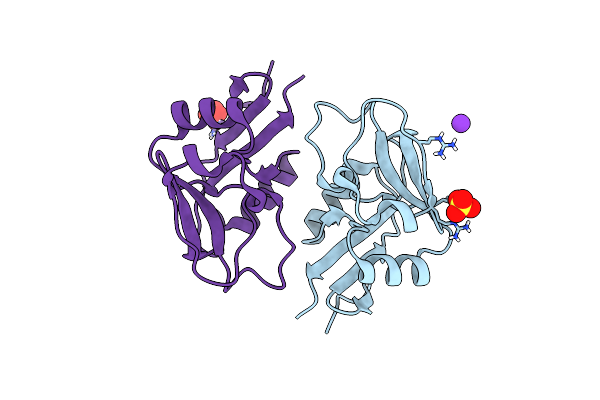 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-03-13 Classification: STRUCTURAL PROTEIN Ligands: SO4, K |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: MEMBRANE PROTEIN Ligands: PIO |
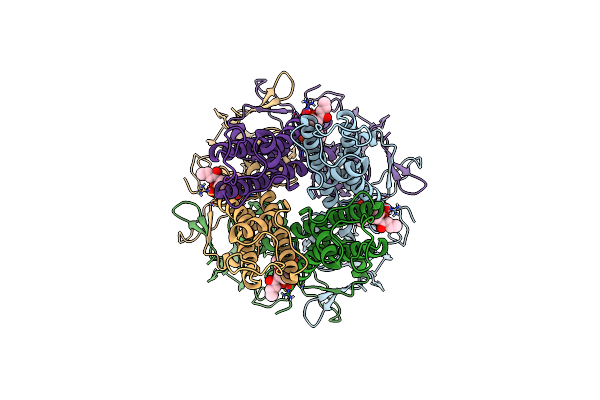 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: MEMBRANE PROTEIN Ligands: PIO |
 |
 |
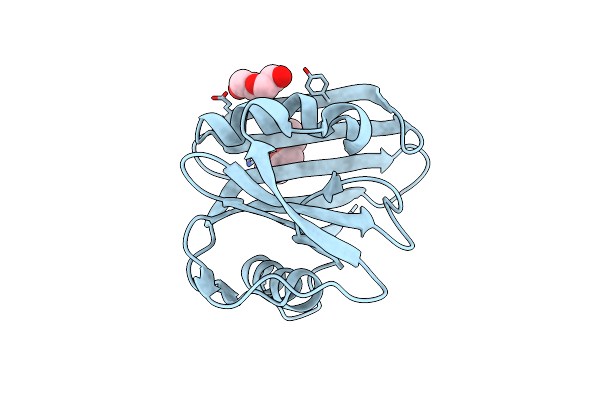
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-08-09 Classification: STRUCTURAL PROTEIN |
 |
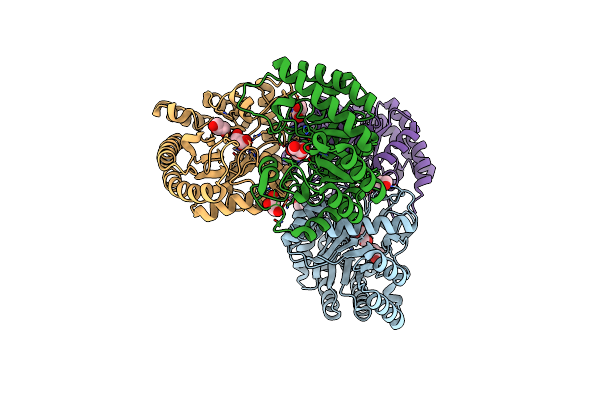
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: PG4, PEG |
 |
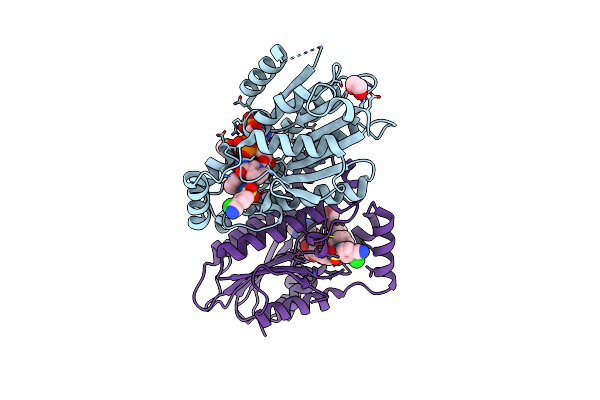
Organism: Ruminiclostridium cellulolyticum (strain atcc 35319 / dsm 5812 / jcm 6584 / h10)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-24 Classification: ISOMERASE Ligands: MN, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, H4H, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, H4E, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, H4T, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, H5B, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, H6E, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: NAP, H5E |

