Search Count: 35
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: EVB, BDF |
 |
Organism: Enterobacteria phage l1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-12-04 Classification: SUGAR BINDING PROTEIN Ligands: NDG, FWQ |
 |
Organism: Escherichia phage johannlburckhardt
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-20 Classification: VIRAL PROTEIN |
 |
Organism: Salmonella, Bacteriophage sp.
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-11-06 Classification: RNA BINDING PROTEIN Ligands: GOL, ATP, MG, NA |
 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Double Mutant Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CA, A1H0D, LI |
 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Single Mutant Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CA, A1H0D |
 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Wild Type Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: A1H0D, CA, LI |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: BDF, EVB, GOL |
 |
The Rsl-D32N - Sulfonato-Calix[8]Arene Complex, I23 Form, Citrate Ph 4.0, Obtained By Cross-Seeding
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: EVB, BDF, GOL |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: GOL, BDF, EVB |
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Homocysteine Bound.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
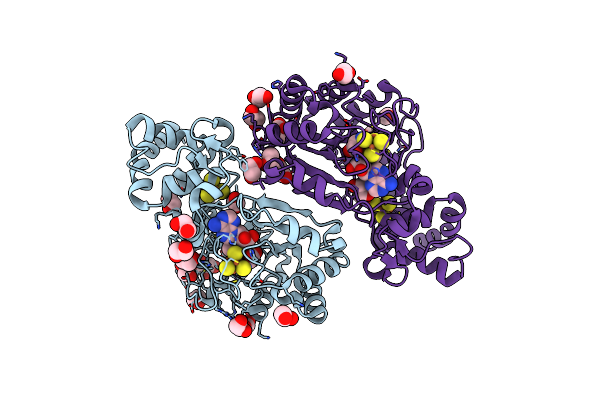
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
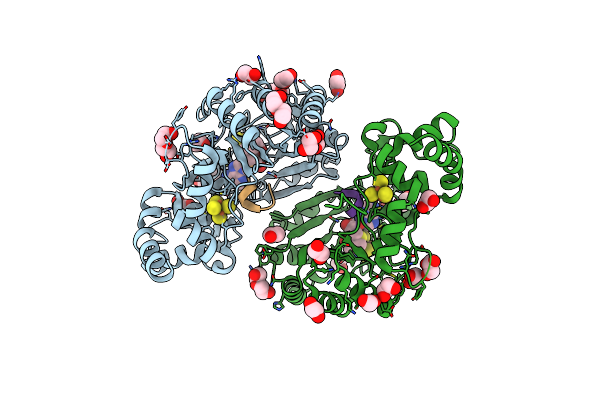
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Methionine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAM, EDO, GOL |
 |
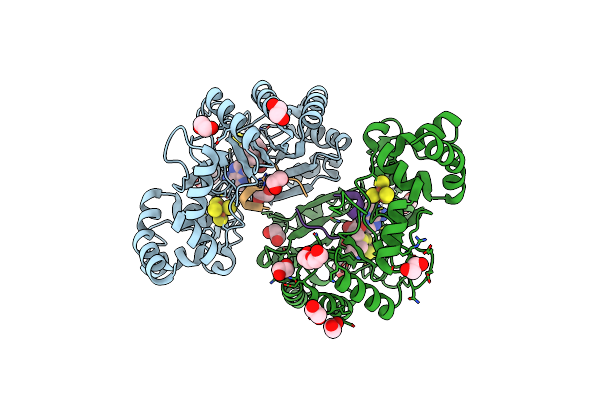
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG, GOL |
 |
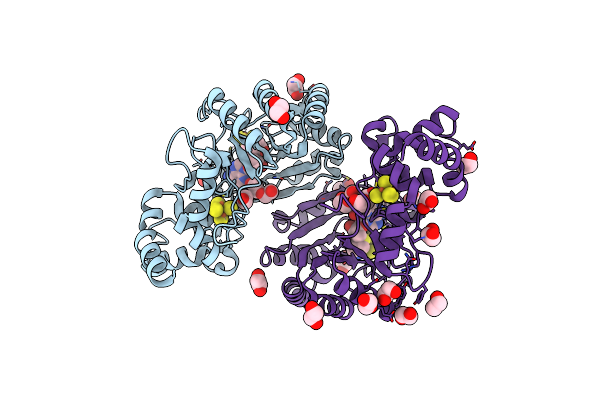
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 6 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee D210A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Persulfurated Cysteine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Structure Of Protease1 From Pyrococcus Horikoshii In Space Group 19 With A Hexamer In The Asymmetric Unit
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-03-30 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-20 Classification: HYDROLASE Ligands: SO4 |
 |
Structure Of The Pl6 Family Alginate Lyase Patl3640 From Pseudoalteromonas Atlantica T6C
Organism: Pseudoalteromonas atlantica (strain t6c / atcc baa-1087)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2021-07-28 Classification: LYASE Ligands: GOL |
 |
Structure Of The Pl6 Family Chondroitinase B From Pseudopedobacter Saltans, Pedsa3807
Organism: Pseudopedobacter saltans (strain atcc 51119 / dsm 12145 / jcm 21818 / lmg 10337 / nbrc 100064 / ncimb 13643)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-07-28 Classification: LYASE |

