Search Count: 57
 |
Crystal Structure Of Lmrr Variant V15Ay With Val15 Replaced By 3-Aminotyrosine
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA BINDING PROTEIN |
 |
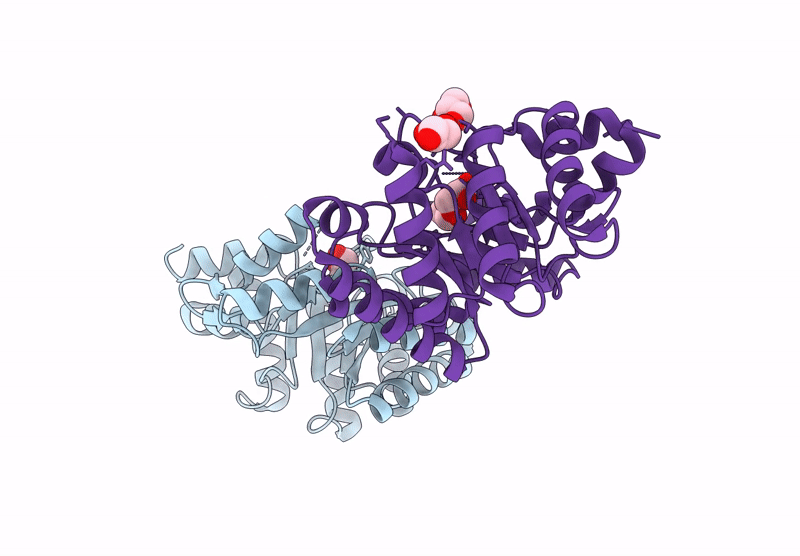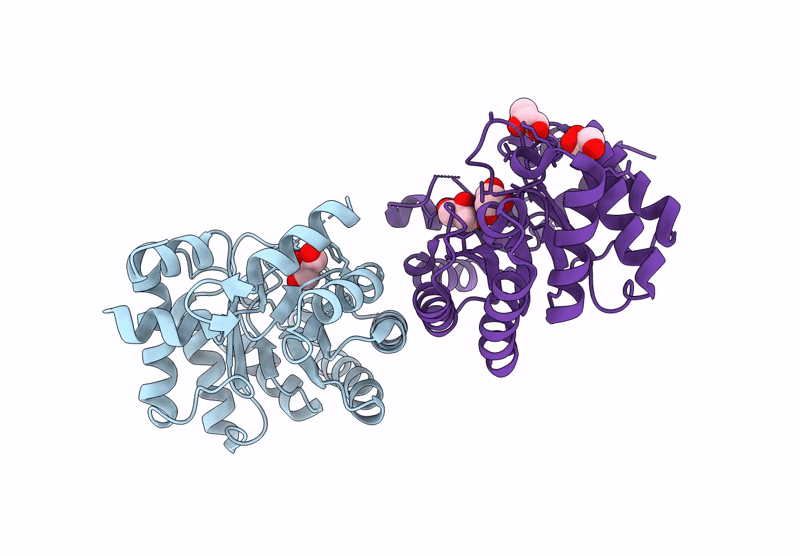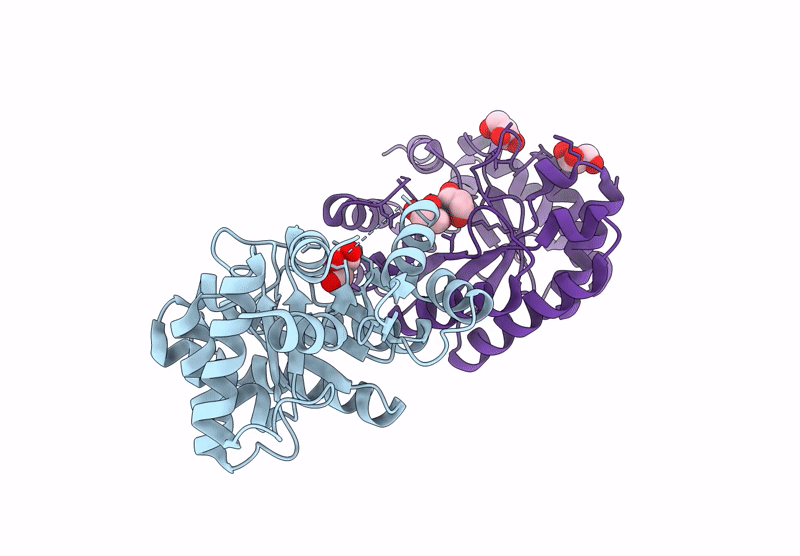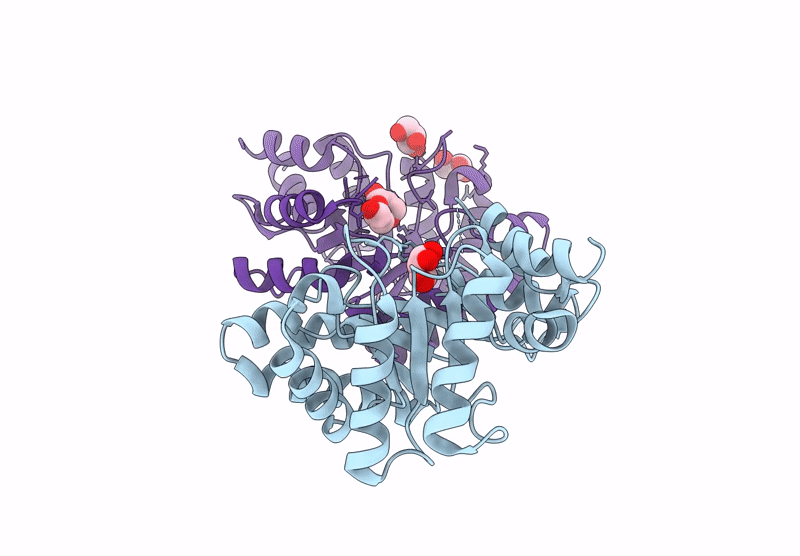
Crystal Structure Of Lmrr Variant V15Ay-Rnyw With Val15 Replaced By 3-Aminotyrosine And Evolved As Friedel-Crafts Alkylase
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA BINDING PROTEIN Ligands: NO3 |
 |
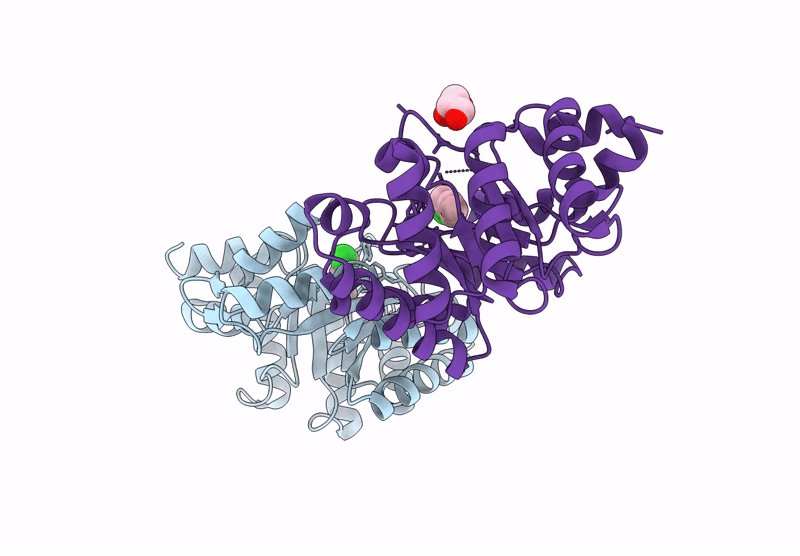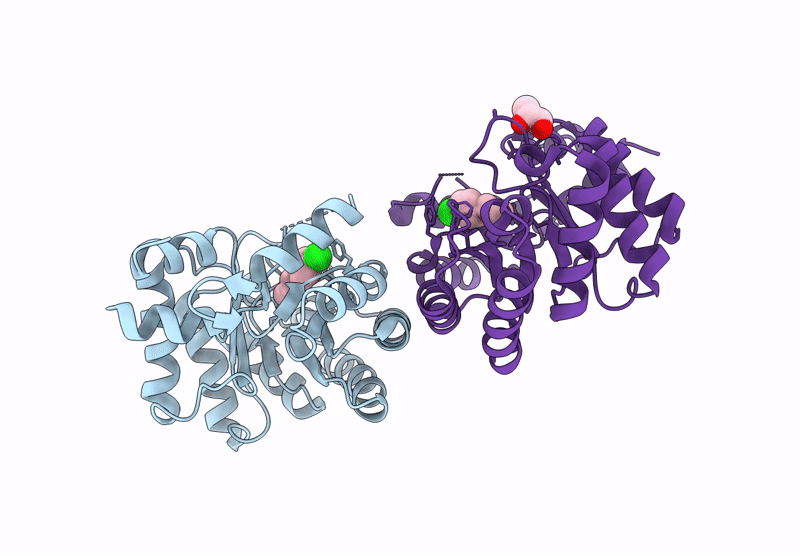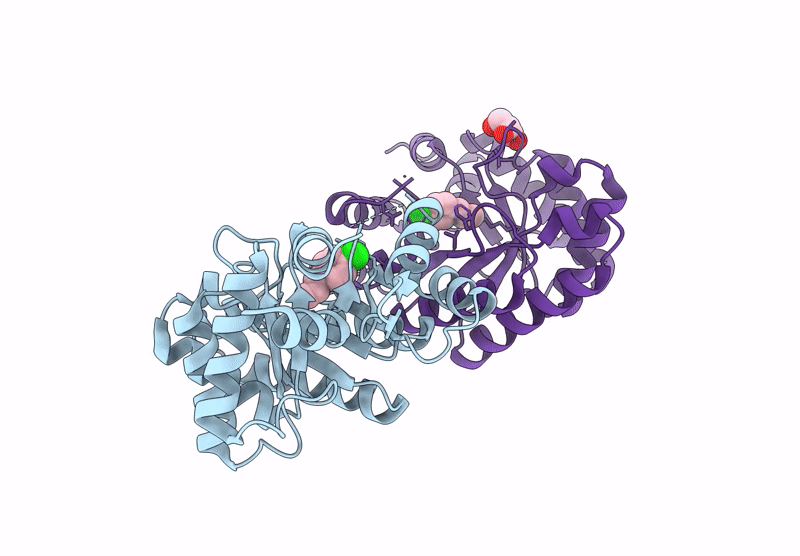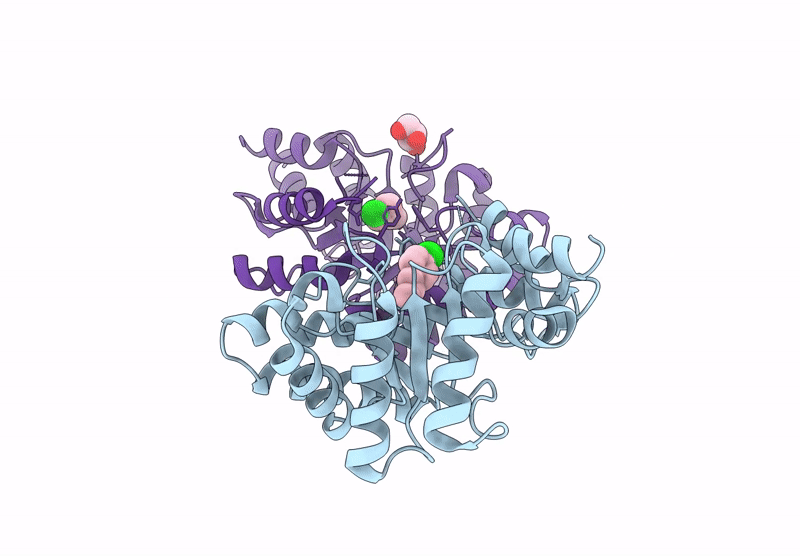
Re-Engineered Peroxygenase Variant Of 2-Deoxy-D-Ribose-5-Phosphate Aldolase In Substrate-Free State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-03-12 Classification: LYASE Ligands: GOL |
 |
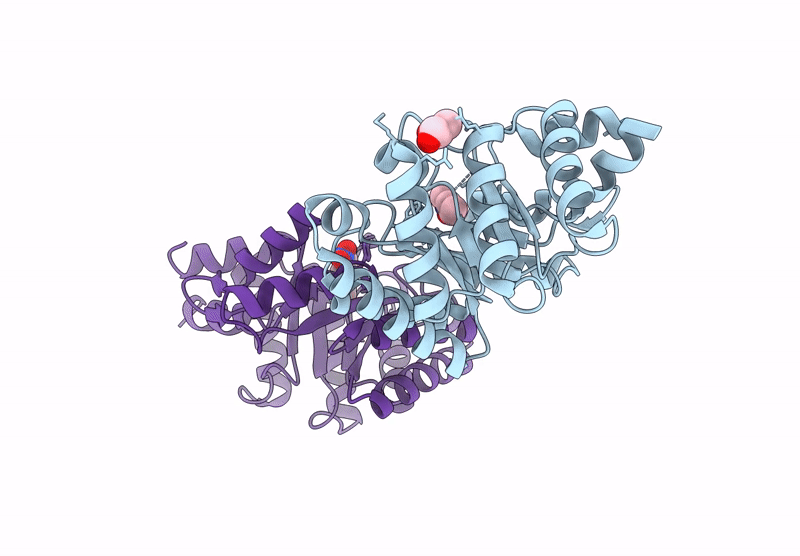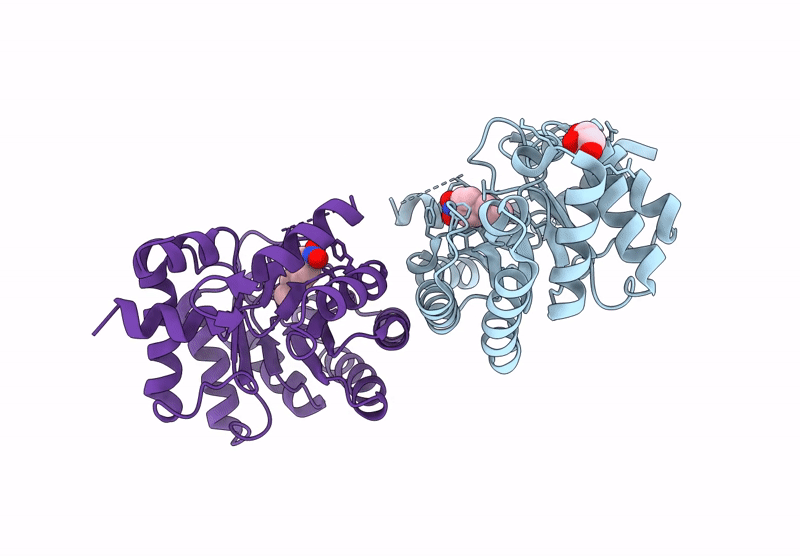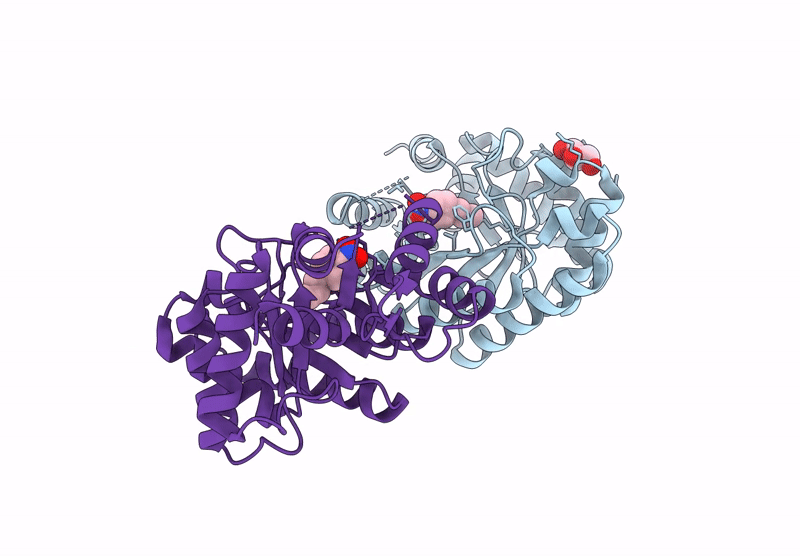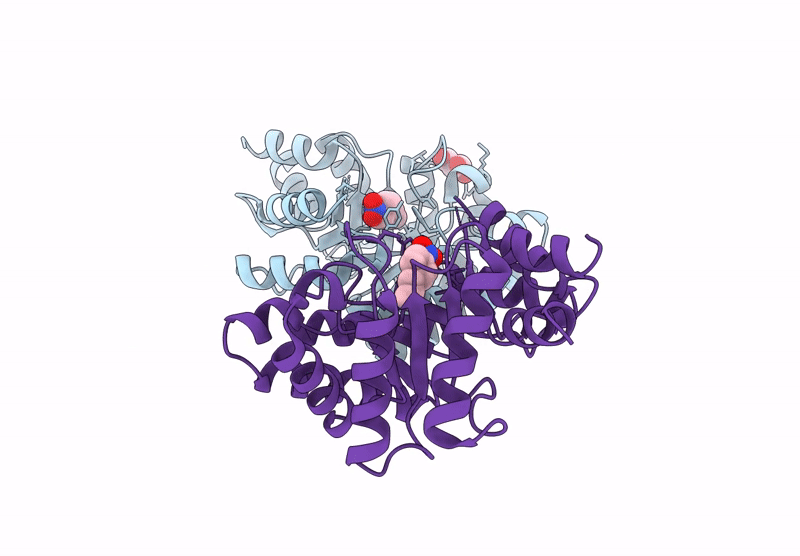
Re-Engineered Peroxygenase Variant Of 2-Deoxy-D-Ribose-5-Phosphate Aldolase, Schiff-Base Complex With 4-Chloro-Cinnamaldehyde
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-03-12 Classification: LYASE Ligands: A1ICA, GOL |
 |
Re-Engineered Peroxygenase Variant Of 2-Deoxy-D-Ribose-5-Phosphate Aldolase, Schiff-Base Complex With 4-Nitro-Cinnamaldehyde
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-03-12 Classification: LYASE Ligands: A1IB9, GOL |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-03-05 Classification: METAL BINDING PROTEIN Ligands: B3P |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-03-05 Classification: METAL BINDING PROTEIN |
 |
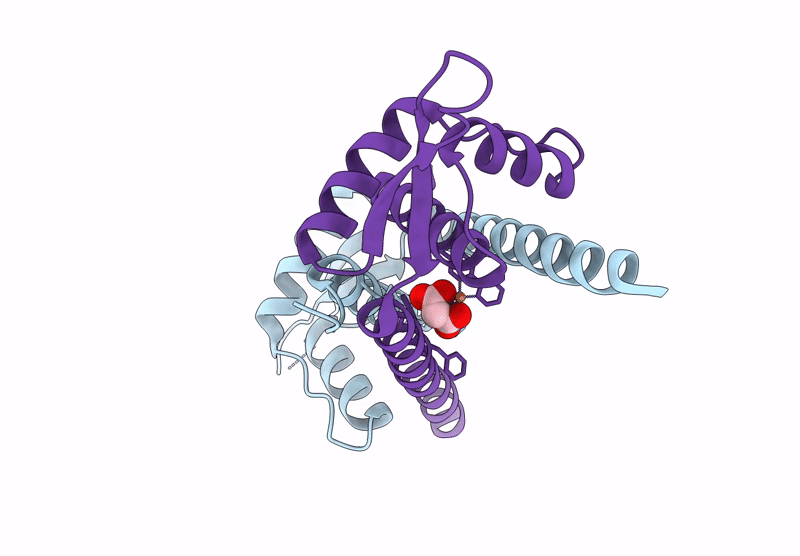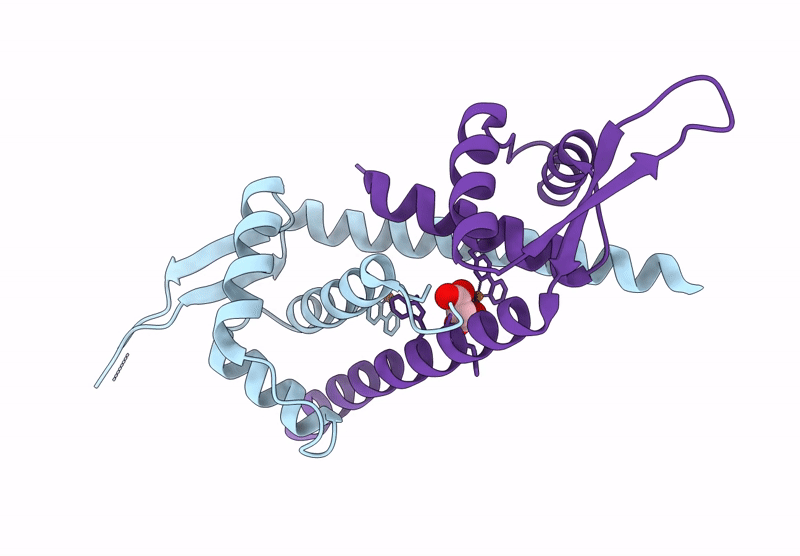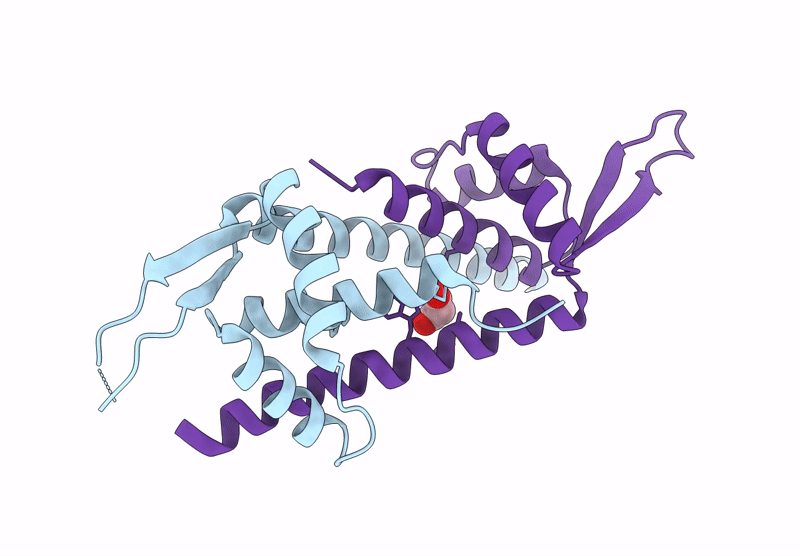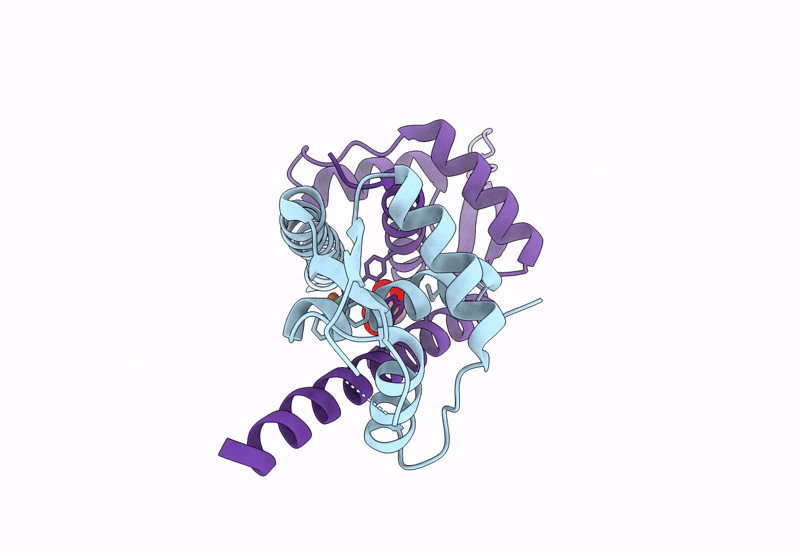
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-03-05 Classification: METAL BINDING PROTEIN |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-03-05 Classification: METAL BINDING PROTEIN Ligands: CU, MLI |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-03-05 Classification: METAL BINDING PROTEIN Ligands: GOL, IMD, CU |
 |
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 1
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
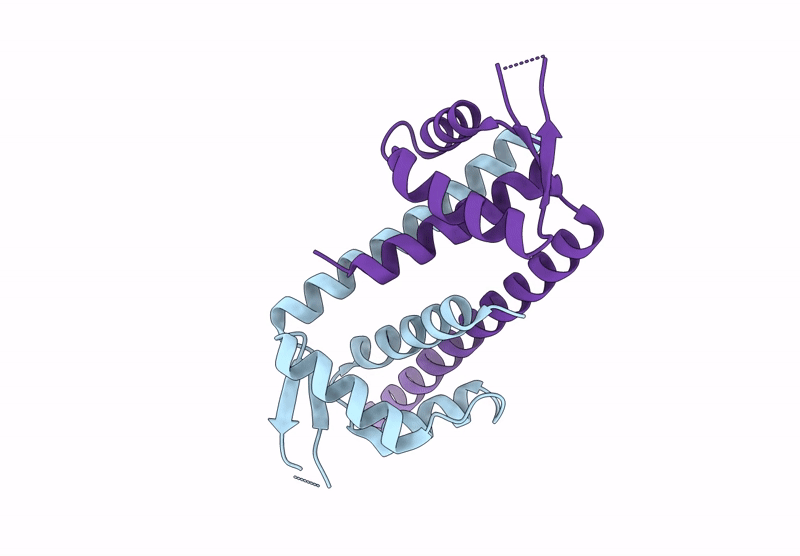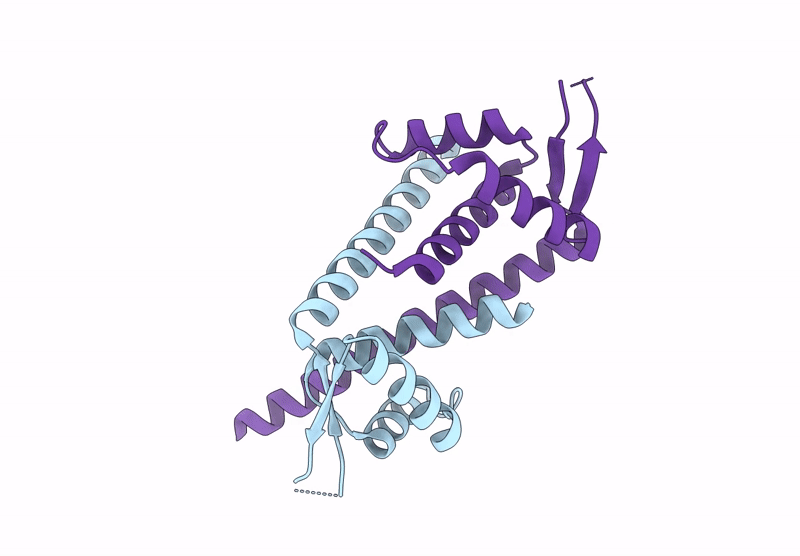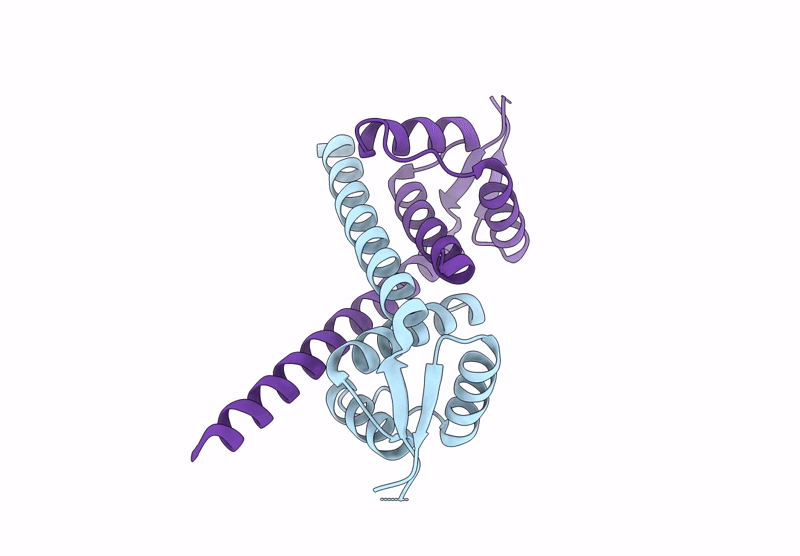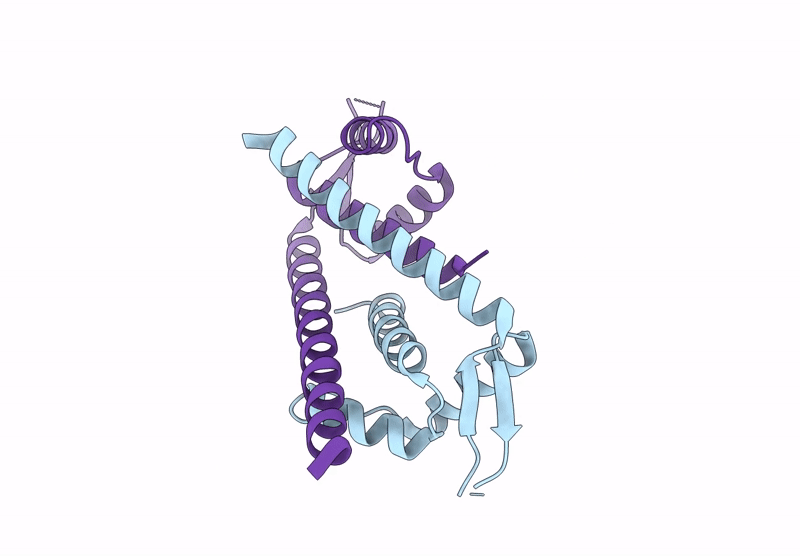
Crystal Structure Of Artificial Enzyme Lmrr_Paf Variant Rmh In Crystal Form 2
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Mutant Aspartase From Bacillus Sp. Ym55-1 In The Closed Loop Conformation
Organism: Bacillus sp. ym55-1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: LYASE Ligands: PGE, NA |
 |
Crystal Structure Of Mutant Aspartase From Caenibacillus Caldisaponilyticus In The Closed Loop Conformation
Organism: Caenibacillus caldisaponilyticus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-15 Classification: LYASE |
 |
Crystal Structure Of Lmrr With V15 Replaced By Unnatural Amino Acid 4-Mercaptophenylalanine, Apo
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: HOH |
 |
Crystal Structure Of Lmrr With V15 Replaced By Unnatural Amino Acid 4-Mercaptophenylalanine, Au(I) Bound
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: AU |
 |
Crystal Structure Of Ramr With Tyr59 Replaced With Para-Boronophenylalanine (Boronate Form)
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-12-25 Classification: DNA BINDING PROTEIN |
 |
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-05-01 Classification: TRANSCRIPTION |
 |
Engineered Lmrr Carrying A Cyclic Boronate Ester Formed Between Tris And P-Boronophenylalanine At Position 89
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-05-01 Classification: TRANSCRIPTION Ligands: GOL |

