Search Count: 26
 |
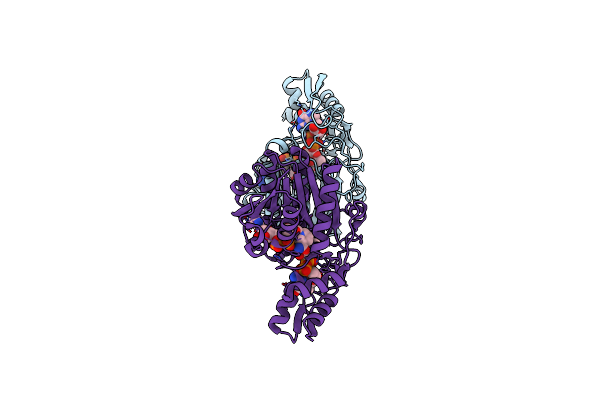
Crystal Structure Of Udp-Galactose 4-Epimerase From Pyrococcus Horikoshii With Bound Nad
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: NAD |
 |
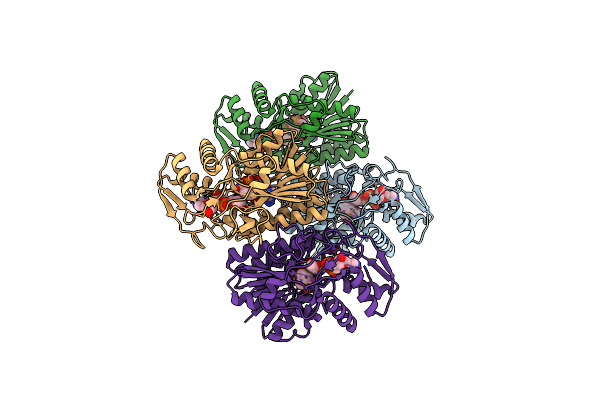
Crystal Structure Of Udp-Galactose 4-Epimerase From Pyrococcus Horikoshii With Bound Nad And Gdp-L-Fucose
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: NAD, GFB |
 |
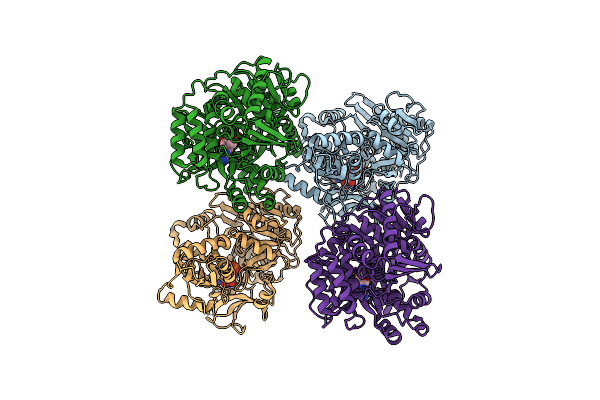
Crystal Structure Of Udp-Galactose 4-Epimerase From Pyrococcus Horikoshii Containing Y145F Mutation And With Bound Nad And Gdp-L-Fucose
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: NAD, GFB |
 |
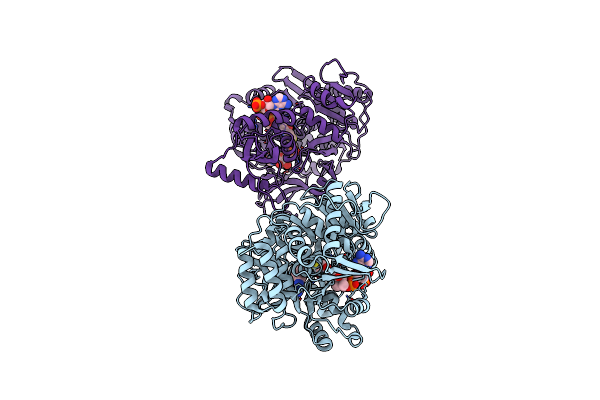
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: AMP, 6R9 |
 |
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Coenzyme A And Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: 6R9, COA |
 |
Organism: Faecalibaculum rodentium
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: TRS |
 |
Organism: Jeotgalibaca ciconiae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: TRS |
 |
Organism: Lactococcus lactis subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-01 Classification: DNA BINDING PROTEIN Ligands: PHN, CU |
 |
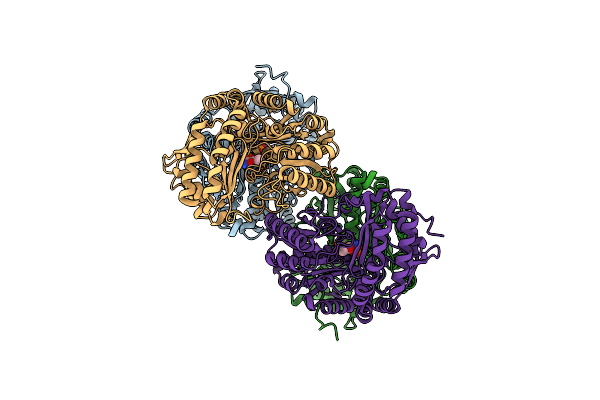
Amine Transaminase Crystal Structure From An Uncultivated Pseudomonas Species In The Plp-Bound (Internal Aldimine) Form
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: PLP |
 |
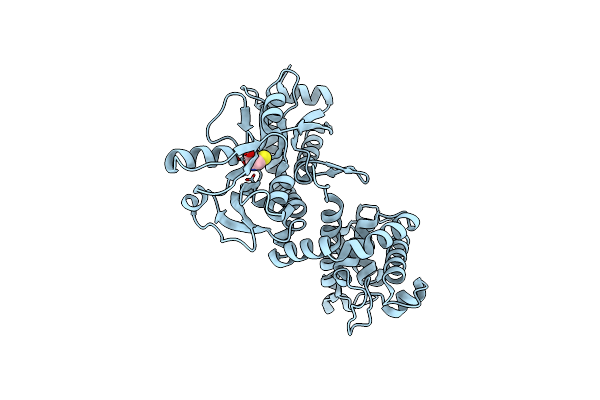
Amine Transaminase Crystal Structure From An Uncultivated Pseudomonas Species In The Pmp-Bound Form
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-03-22 Classification: TRANSFERASE Ligands: PMP |
 |
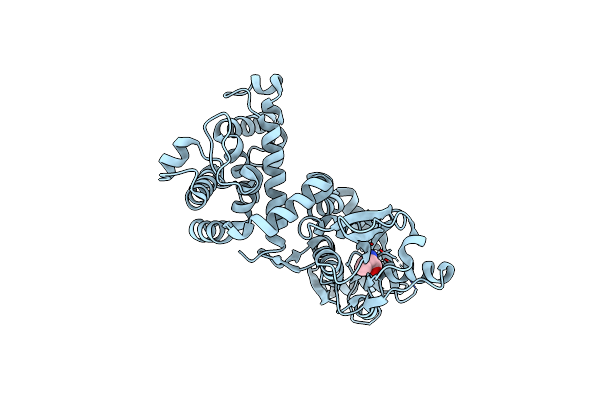
Crystal Structure Of Mltf From Pseudomonas Aeruginosa Complexed With Cysteine
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2015-03-18 Classification: LYASE Ligands: CYS, CL |
 |
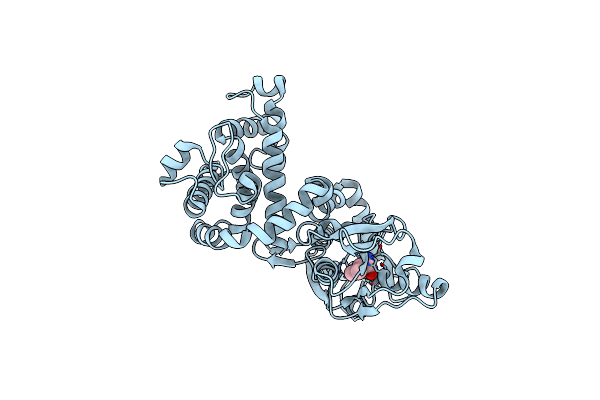
Crystal Structure Of Mltf From Pseudomonas Aeruginosa Complexed With Valine
Organism: Pseudomonas aeruginosa padk2_cf510
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-03-18 Classification: LYASE Ligands: VAL |
 |
Crystal Structure Of Mltf From Pseudomonas Aeruginosa Complexed With Leucine
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2015-03-18 Classification: LYASE Ligands: LEU, CL |
 |
Crystal Structure Of Mltf From Pseudomonas Aeruginosa Complexed With Isoleucine
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2015-03-18 Classification: LYASE Ligands: ILE |
 |
Crystal Structure Of Mltf From Pseudomonas Aeruginosa Complexed With Bulgecin And Muropeptide
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-03-18 Classification: LYASE Ligands: BLG, MG, CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2015-03-18 Classification: LYASE Ligands: MG, CL |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-11-06 Classification: OXIDOREDUCTASE Ligands: FAD, NA, CL, PG4 |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-11-06 Classification: OXIDOREDUCTASE Ligands: FAD, NA, PG4, ANB |
 |
Organism: Clostridium tetanomorphum
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2012-05-02 Classification: LYASE Ligands: MG, GOL, CL |
 |
Organism: Clostridium tetanomorphum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-05-02 Classification: LYASE Ligands: GOL, ROP, MG, CL |

