Search Count: 55
 |
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: MG |
 |
Organism: Zobellia galactanivorans
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: IMD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-10-04 Classification: TRANSCRIPTION Ligands: U52 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-10-04 Classification: TRANSCRIPTION Ligands: RC0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-10-04 Classification: TRANSCRIPTION Ligands: TJ3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-07 Classification: LYASE Ligands: PBG, ZN, GOL |
 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2016-10-12 Classification: LYASE Ligands: ZN |
 |
X-Ray Structure Of Human Recombinant 5-Aminolaevulinic Acid Dehydratase (Hralad).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-01-27 Classification: LYASE Ligands: ZN |
 |
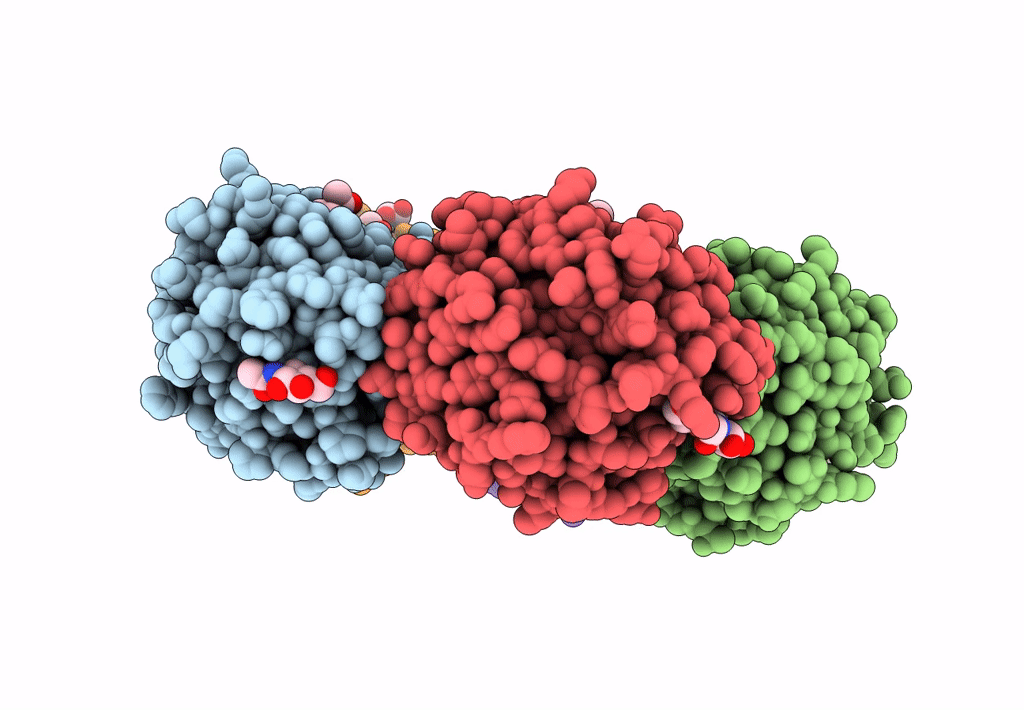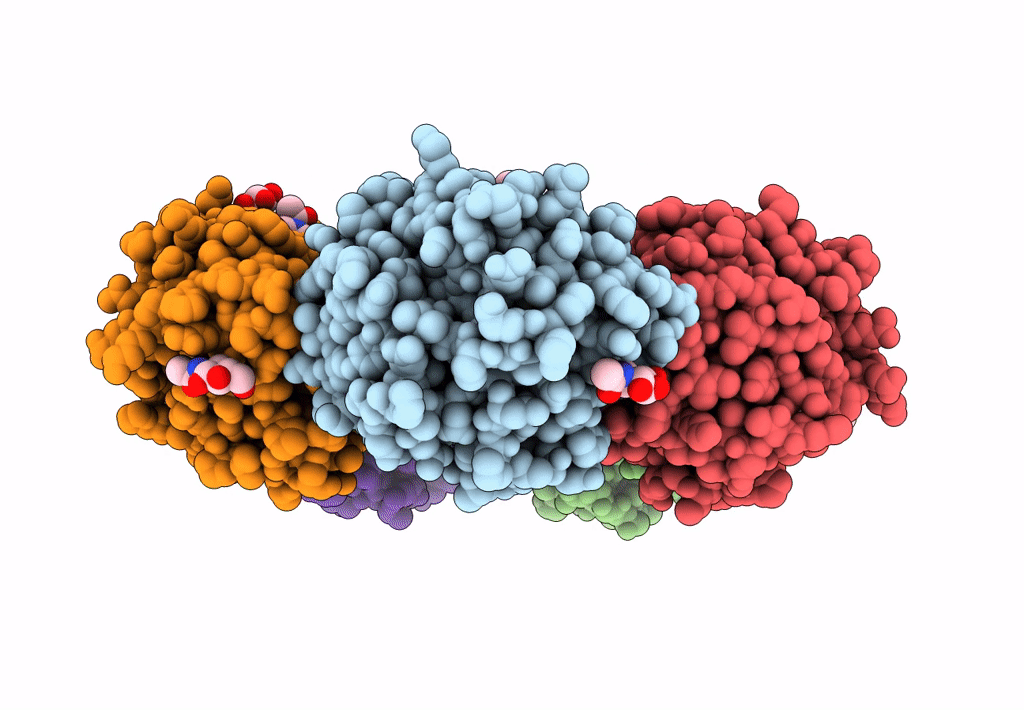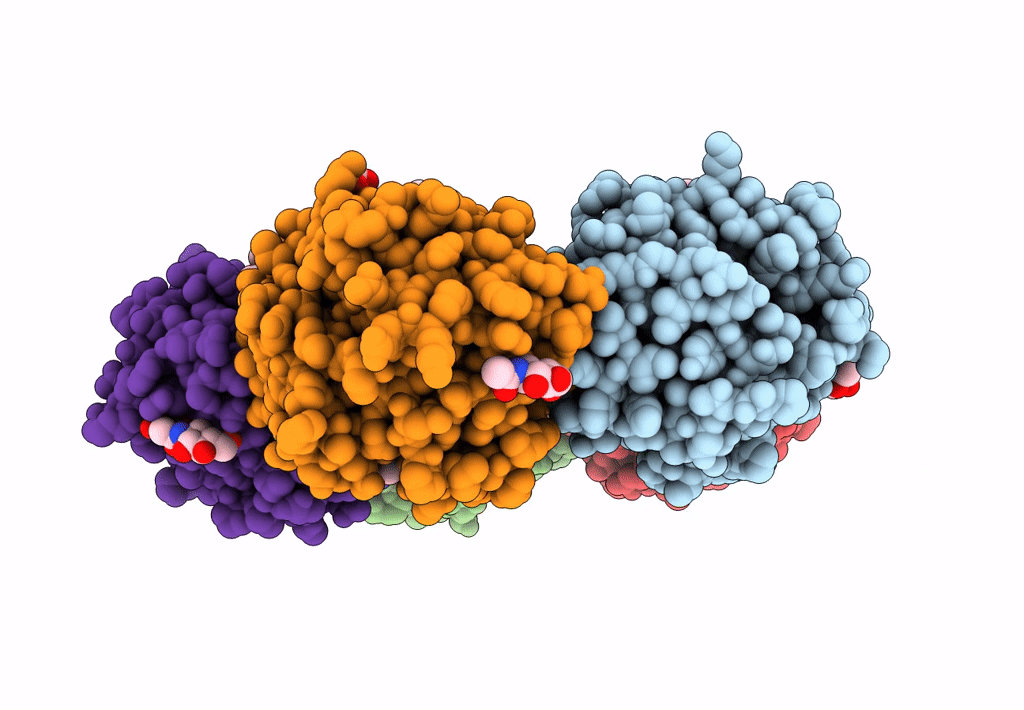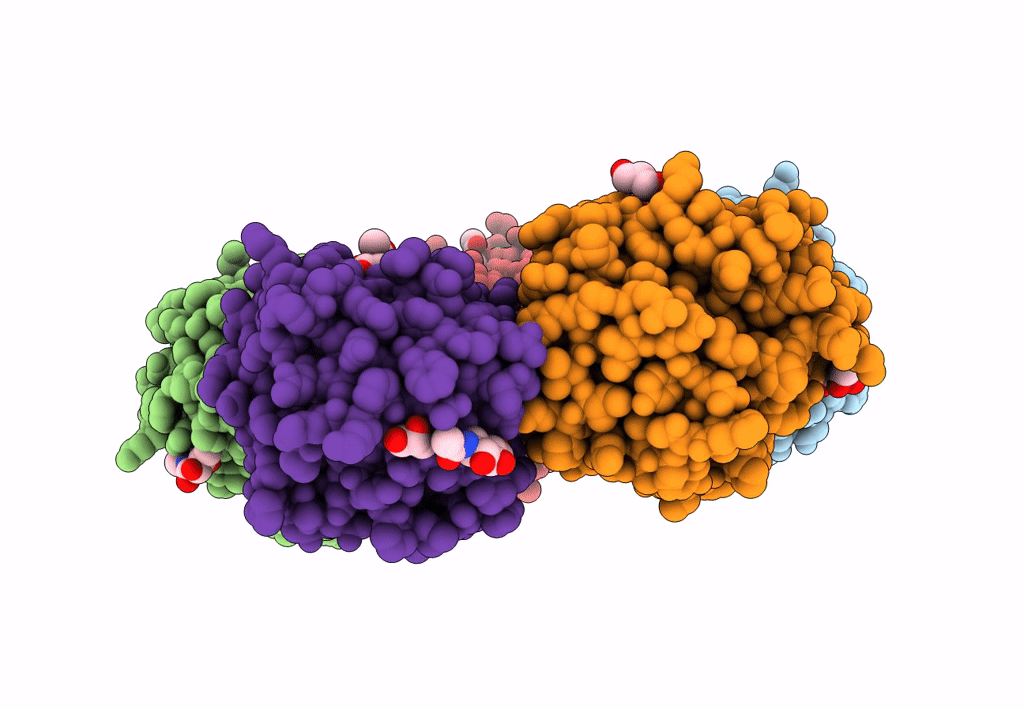
The X-Ray Structure Of Octameric Human Native 5-Aminolaevulinic Acid Dehydratase.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2016-01-27 Classification: LYASE Ligands: ZN, SO4, DAV |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-07-10 Classification: SUGAR BINDING PROTEIN Ligands: CA, N8P, NAG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-06-19 Classification: SUGAR BINDING PROTEIN Ligands: NAG, CA, N7P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-06-19 Classification: SUGAR BINDING PROTEIN Ligands: CA, NAG, GHE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-06-19 Classification: SUGAR BINDING PROTEIN Ligands: ACT, GHE, CD, NAG, SIA |
 |
Crystal Structure Of The C-Terminal Domain Of Nuclear Pore Complex Component Nup116 From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-08-04 Classification: PROTEIN TRANSPORT Ligands: GOL |
 |
Crystal Structure Of The C-Terminal Domain From The Nuclear Pore Complex Component Nup133 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-26 Classification: PROTEIN TRANSPORT Ligands: GOL |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae In The Hexagonal, P61 Space Group
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Pseudomonas fluorescens pf-5
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2009-10-20 Classification: TRANSPORT PROTEIN Ligands: LDA |
 |
Crystal Structure Of The Second Phr Domain Of Mouse Myc-Binding Protein 2 (Mycbp-2)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-07-21 Classification: LIGASE Ligands: DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2009-07-07 Classification: ONCOPROTEIN Ligands: MG, CL, ATP |

