Search Count: 25
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: NAG |
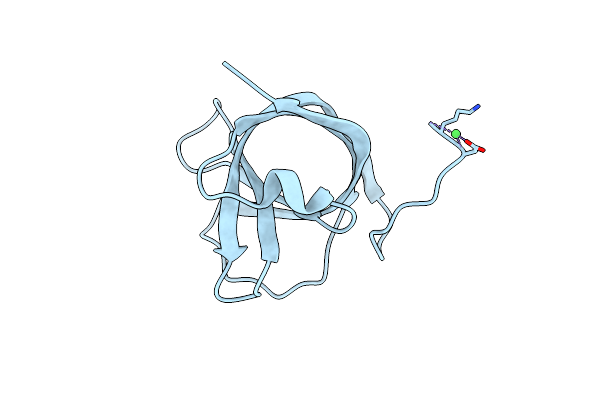 |
Matrix-Metallopeptidase Inhibitor Potempin A (Pota) From Tannerella Forsythia
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: NI |
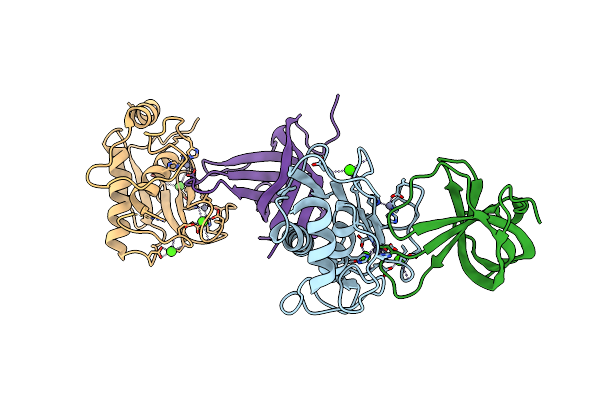 |
Potempin A (Pota) From Tannerella Forsythia In Complex With The Catalytic Domain Of Human Mmp-12
Organism: Homo sapiens, Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: CA, ZN |
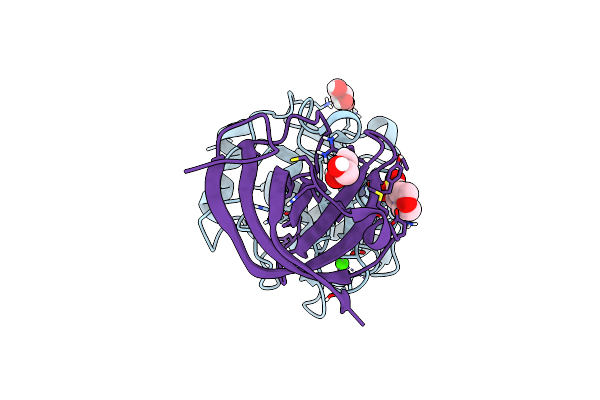 |
Matrix-Metallopeptidase Inhibitor Potempin A (Pota) From Tannerella Forsythia In Complex With T. Forsythia Karilysin.
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: ZN, CA, MES, GOL |
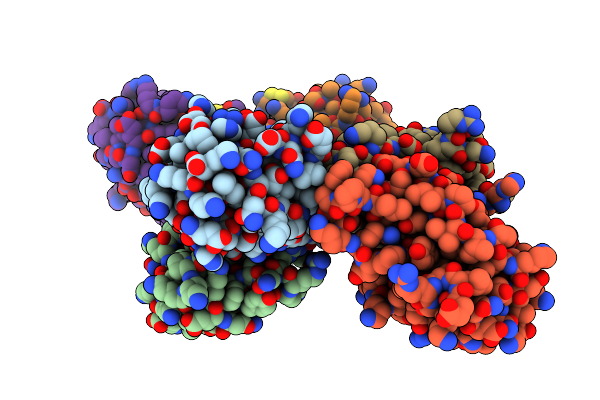 |
Structure Of Tannerella Forsythia Selenomethionine-Derivatized Potempin D Mutant I53M
Organism: Tannerella forsythia (strain atcc 43037 / jcm 10827 / ccug 21028 a / kctc 5666 / fdc 338)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: EDO |
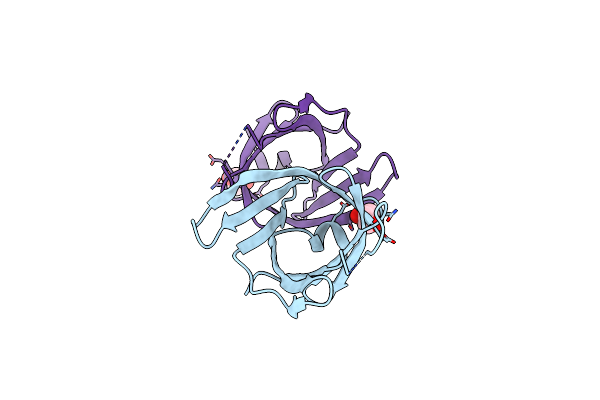 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: GOL |
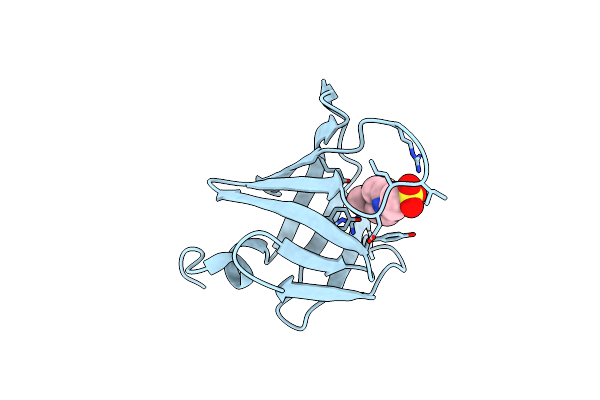 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: EPE |
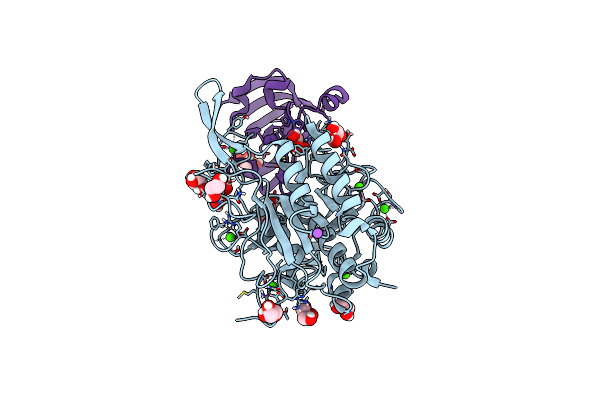 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR |
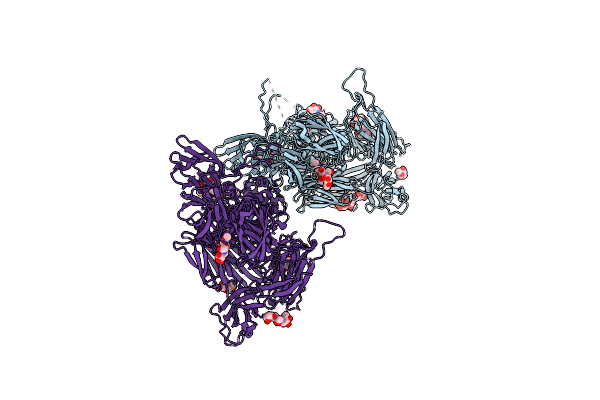 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 2022-04-20 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-08-04 Classification: HYDROLASE Ligands: V7Q, EDO, ACT, ZN, CA, CL |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-06 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Porphyromonas gingivalis w83
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2016-04-06 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of Dionain-1, The Major Endopeptidase In The Venus Flytrap Digestive Juice
Organism: Dionaea muscipula
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-12-09 Classification: HYDROLASE Ligands: E64, PO4 |

